Section 4.6 — Generalized linear models#
This notebook contains the code examples from Section 4.6 Generalized linear models from the No Bullshit Guide to Statistics.
Notebook setup#
# Ensure required Python modules are installed
%pip install --quiet numpy scipy seaborn pandas statsmodels ministats
Note: you may need to restart the kernel to use updated packages.
# Load Python modules
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
# Figures setup
plt.clf() # needed otherwise `sns.set_theme` doesn't work
sns.set_theme(
context="paper",
style="whitegrid",
palette="colorblind",
rc={"font.family": "serif",
"font.serif": ["Palatino", "DejaVu Serif", "serif"],
"figure.figsize": (5,3)},
)
%config InlineBackend.figure_format = "retina"
<Figure size 640x480 with 0 Axes>
# Simple float __repr__
if int(np.__version__.split(".")[0]) >= 2:
np.set_printoptions(legacy='1.25')
# Download datasets/ directory if necessary
from ministats import ensure_datasets
ensure_datasets()
datasets/ directory already exists.
Definitions#
Probability representations and link functions#
Odds#
0.5/(1-0.5), 0.9/(1-0.9), 0.2/(1-0.2)
(1.0, 9.000000000000002, 0.25)
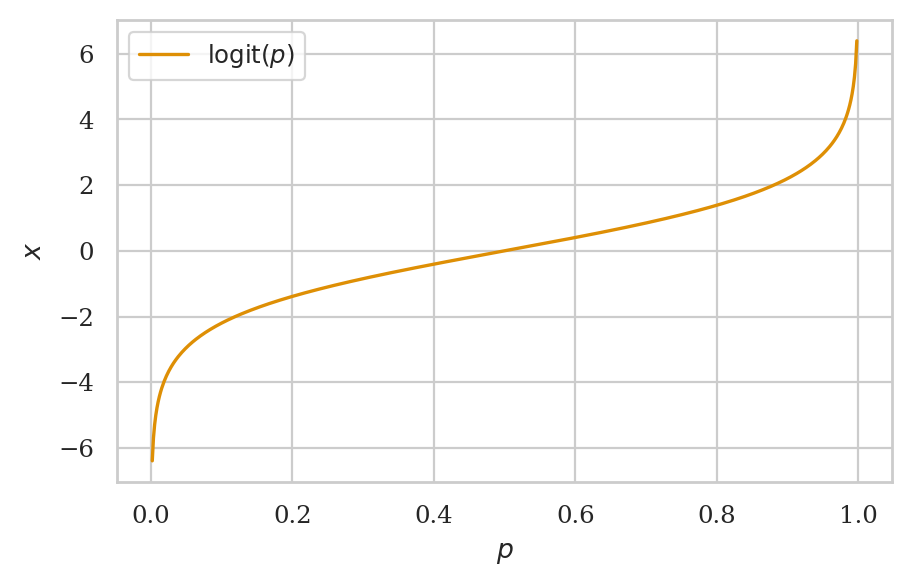
Log-odds#
np.log(0.5/(1-0.5)), np.log(0.9/(1-0.9)), np.log(0.2/(1-0.2))
(0.0, 2.1972245773362196, -1.3862943611198906)
The logit function#
def logit(p):
x = np.log(p / (1-p))
return x
# ALT. import the function from `scipy.special`
from scipy.special import logit
logit(0.5), logit(0.9), logit(0.2)
(0.0, 2.1972245773362196, -1.3862943611198906)
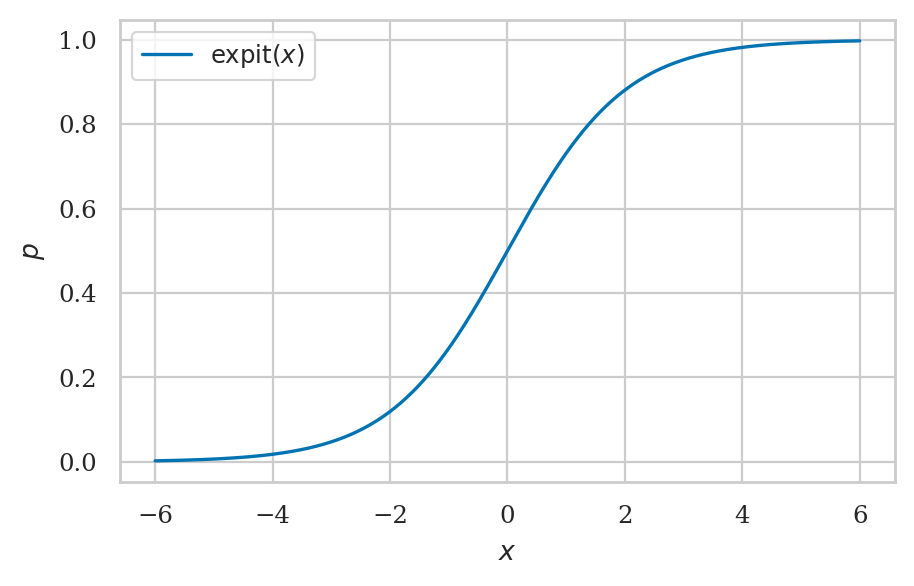
The logistic function#
def expit(x):
p = 1 / (1 + np.exp(-x))
return p
# ALT. import the function from `scipy.special`
from scipy.special import expit
expit(0), expit(2), expit(-2)
(0.5, 0.8807970779778823, 0.11920292202211755)
The logistic and logit functions are inverses#
expit(logit(0.2))
0.2
logit(expit(3))
3.000000000000003
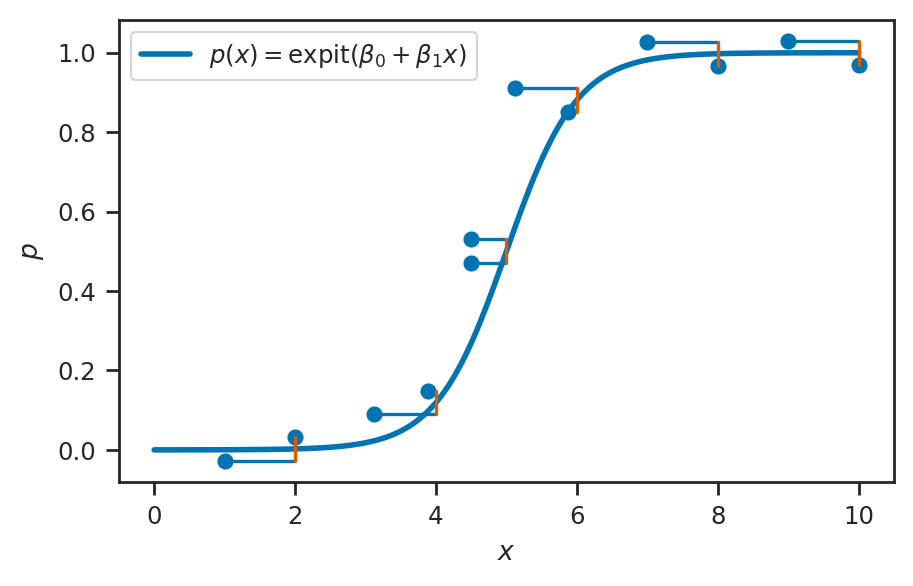
Logistic regression#
TODO FORMULA
from scipy.stats import bernoulli
from scipy.special import expit
# Define the logistic regression model function
def expit_model(x):
p = expit(-10 + 2*x)
return p
xlims = [0, 10]
stem_half_width = 0.03
with sns.axes_style("ticks"):
fig, ax = plt.subplots(figsize=(5, 3))
# Plot the logistic regression model
xs = np.linspace(xlims[0], xlims[1], 200)
ps = expit_model(xs)
sns.lineplot(x=xs, y=ps, ax=ax, label=r"$p(x) = \mathrm{expit}(\beta_0 + \beta_1x)$", linewidth=2)
# Plot Bernoulli distributions at specified x positions
x_positions = [2,4,5,6,8,10]
for x_pos in x_positions:
p_pos = expit_model(x_pos)
ys = [0,1]
pmf = bernoulli(p=p_pos).pmf(ys)
ys_plot = [p_pos-stem_half_width, p_pos+stem_half_width]
ax.stem(ys_plot, x_pos - pmf, bottom=x_pos, orientation='horizontal')
# Figure setup
ax.set_xlabel("$x$")
ax.set_ylabel("$p$")
ax.legend(loc="upper left")
expit(-6)
0.0024726231566347743
expit(10)
0.9999546021312976
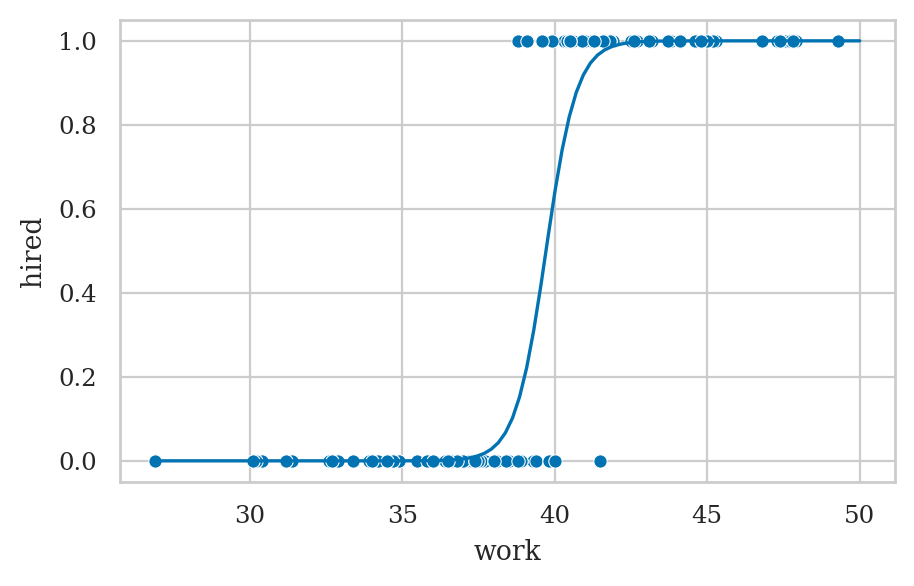
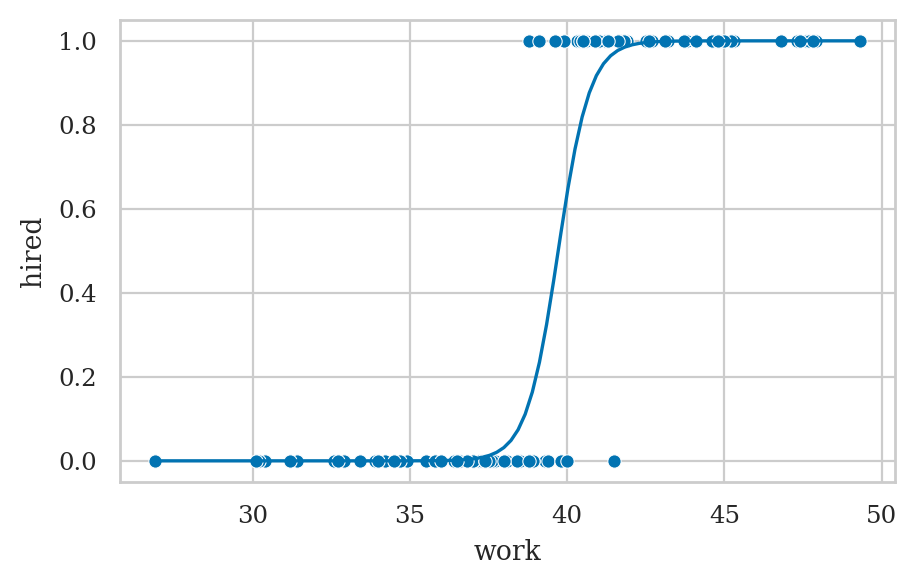
Example 1: hiring student interns#
interns = pd.read_csv("datasets/interns.csv")
print(interns.shape)
interns.head(3)
(100, 2)
| work | hired | |
|---|---|---|
| 0 | 42.5 | 1 |
| 1 | 39.3 | 0 |
| 2 | 43.2 | 1 |
import statsmodels.formula.api as smf
lr1 = smf.logit("hired ~ 1 + work", data=interns).fit()
print(lr1.params)
Optimization terminated successfully.
Current function value: 0.138101
Iterations 10
Intercept -78.693205
work 1.981458
dtype: float64
lr1.summary()
| Dep. Variable: | hired | No. Observations: | 100 |
|---|---|---|---|
| Model: | Logit | Df Residuals: | 98 |
| Method: | MLE | Df Model: | 1 |
| Date: | Wed, 04 Mar 2026 | Pseudo R-squ.: | 0.8005 |
| Time: | 22:41:14 | Log-Likelihood: | -13.810 |
| converged: | True | LL-Null: | -69.235 |
| Covariance Type: | nonrobust | LLR p-value: | 6.385e-26 |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -78.6932 | 19.851 | -3.964 | 0.000 | -117.600 | -39.787 |
| work | 1.9815 | 0.500 | 3.959 | 0.000 | 1.001 | 2.962 |
Possibly complete quasi-separation: A fraction 0.32 of observations can be
perfectly predicted. This might indicate that there is complete
quasi-separation. In this case some parameters will not be identified.
ax = sns.scatterplot(data=interns, x="work", y="hired")
wgrid = np.linspace(27, 50, 100)
hired_preds = lr1.predict({"work": wgrid})
sns.lineplot(x=wgrid, y=hired_preds, ax=ax);
lr1.summary()
| Dep. Variable: | hired | No. Observations: | 100 |
|---|---|---|---|
| Model: | Logit | Df Residuals: | 98 |
| Method: | MLE | Df Model: | 1 |
| Date: | Wed, 04 Mar 2026 | Pseudo R-squ.: | 0.8005 |
| Time: | 22:41:15 | Log-Likelihood: | -13.810 |
| converged: | True | LL-Null: | -69.235 |
| Covariance Type: | nonrobust | LLR p-value: | 6.385e-26 |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -78.6932 | 19.851 | -3.964 | 0.000 | -117.600 | -39.787 |
| work | 1.9815 | 0.500 | 3.959 | 0.000 | 1.001 | 2.962 |
Possibly complete quasi-separation: A fraction 0.32 of observations can be
perfectly predicted. This might indicate that there is complete
quasi-separation. In this case some parameters will not be identified.
Interpreting the model parameters#
Parameters as changes in the log-odds#
lr1.params["work"]
1.9814577697476643
Parameters as ratios of odds#
np.exp(lr1.params["work"])
7.25330893626569
Differences in probabilities#
What is the marginal effect of the predictor work
for an intern who invests 40 hours of effort?
# using `statsmodels`
lr1.get_margeff(atexog={1:40}).summary_frame()
| dy/dx | Std. Err. | z | Pr(>|z|) | Conf. Int. Low | Cont. Int. Hi. | |
|---|---|---|---|---|---|---|
| work | 0.45783 | 0.112623 | 4.065157 | 0.000048 | 0.237093 | 0.678567 |
lr1.get_margeff(atexog={1:42}).summary_frame()
| dy/dx | Std. Err. | z | Pr(>|z|) | Conf. Int. Low | Cont. Int. Hi. | |
|---|---|---|---|---|---|---|
| work | 0.020949 | 0.021358 | 0.98084 | 0.326672 | -0.020912 | 0.06281 |
# # ALT. manual calculation plugging into derivative of `expit`
# p40 = lr1.predict({"work":40}).item()
# marg_effect_at_40 = p40 * (1 - p40) * lr1.params['work']
# marg_effect_at_40
Prediction#
p42 = lr1.predict({"work":42})[0]
p42
0.9893134055105761
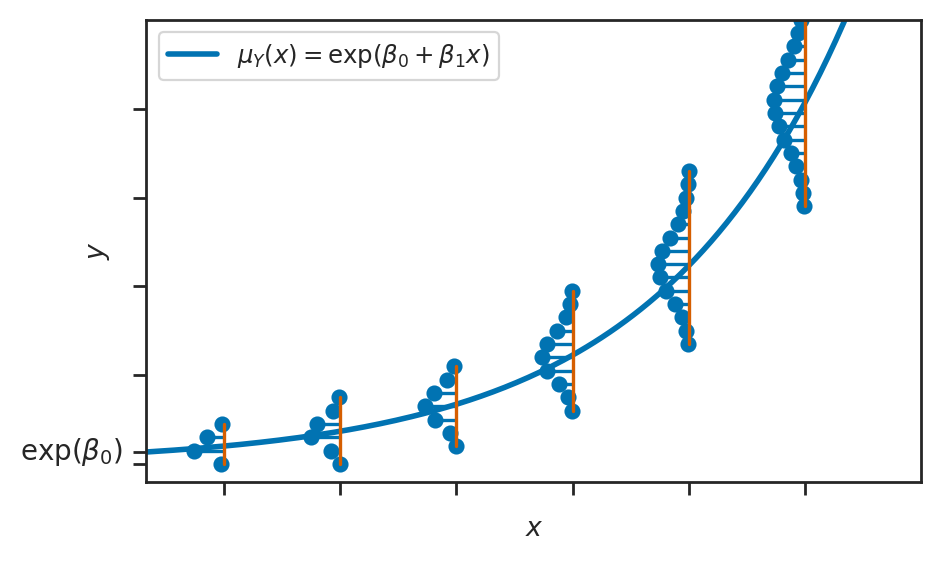
Poisson regression#
from scipy.stats import poisson
# Define the linear model function
def exp_model(x):
lam = np.exp(1 + 0.2*x)
return lam
onepixel = 0.07
xlims = [0, 20]
ylims = [0, 100]
with sns.axes_style("ticks"):
fig, ax = plt.subplots(figsize=(5, 3))
# Plot the linear model
xs = np.linspace(xlims[0], xlims[1], 200)
lams = exp_model(xs)
sns.lineplot(x=xs, y=lams, ax=ax, label=r"$\mu_Y(x) = \exp(\beta_0 + \beta_1x)$", linewidth=2)
# Plot Gaussian distributions at specified x positions and add sigma lines
x_positions = range(2, xlims[1]-1, 3)
for x_pos in x_positions:
lam_pos = exp_model(x_pos)
sigma = np.sqrt(lam_pos)
ys_lower = int(lam_pos-2.5*sigma)
ys_upper = int(lam_pos+3.4*sigma)
ys = np.arange(ys_lower, ys_upper, 3)
pmf = poisson(mu=lam_pos).pmf(ys)
# ax.fill_betweenx(ys, x_pos - 2 * pmf * sigma, x_pos, color="grey", alpha=0.5)
ax.stem(ys, x_pos- 2 * pmf * sigma, bottom=x_pos, orientation='horizontal')
# Draw vertical sigma line and label it on the opposite side of the Gaussian shape
# ax.plot([x_pos+onepixel, x_pos+onepixel], [lam_pos, lam_pos - sigma], "k", lw=1)
# ax.text(x_pos + 0.1, lam_pos - sigma / 2 - 3*onepixel, r"$\sigma$", fontsize=12, va="center")
# y-intercept
ax.text(0 - 0.6, np.exp(1), r"$\exp(\beta_0)$", fontsize=10, va="center", ha="right")
# Set up x-axis
ax.set_xlim(xlims)
ax.set_xlabel("$x$")
ax.set_xticks(range(2, xlims[1], 3))
ax.set_xticklabels([])
# Set up y-axis
ax.set_ylim([ylims[0]-4,ylims[1]])
ax.set_ylabel("$y$")
ax.set_yticks(list(range(ylims[0],ylims[1],20)) + [np.exp(1)] )
ax.set_yticklabels([])
ax.legend(loc="upper left")
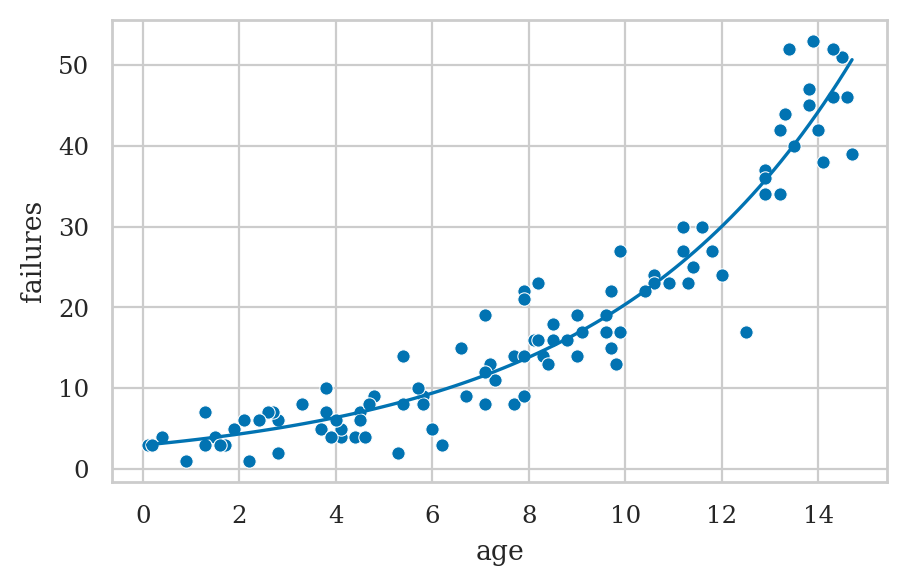
Example 2: hard disk failures over time#
hdisks = pd.read_csv("datasets/hdisks.csv")
hdisks.head(3)
| age | failures | |
|---|---|---|
| 0 | 1.7 | 3 |
| 1 | 14.6 | 46 |
| 2 | 10.9 | 23 |
import statsmodels.formula.api as smf
pr2 = smf.poisson("failures ~ 1 + age", data=hdisks).fit()
pr2.params
Optimization terminated successfully.
Current function value: 2.693129
Iterations 6
Intercept 1.075999
age 0.193828
dtype: float64
pr2.summary()
| Dep. Variable: | failures | No. Observations: | 100 |
|---|---|---|---|
| Model: | Poisson | Df Residuals: | 98 |
| Method: | MLE | Df Model: | 1 |
| Date: | Wed, 04 Mar 2026 | Pseudo R-squ.: | 0.6412 |
| Time: | 22:41:15 | Log-Likelihood: | -269.31 |
| converged: | True | LL-Null: | -750.68 |
| Covariance Type: | nonrobust | LLR p-value: | 2.271e-211 |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 1.0760 | 0.076 | 14.114 | 0.000 | 0.927 | 1.225 |
| age | 0.1938 | 0.007 | 28.603 | 0.000 | 0.181 | 0.207 |
pr2.summary()
| Dep. Variable: | failures | No. Observations: | 100 |
|---|---|---|---|
| Model: | Poisson | Df Residuals: | 98 |
| Method: | MLE | Df Model: | 1 |
| Date: | Wed, 04 Mar 2026 | Pseudo R-squ.: | 0.6412 |
| Time: | 22:41:15 | Log-Likelihood: | -269.31 |
| converged: | True | LL-Null: | -750.68 |
| Covariance Type: | nonrobust | LLR p-value: | 2.271e-211 |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 1.0760 | 0.076 | 14.114 | 0.000 | 0.927 | 1.225 |
| age | 0.1938 | 0.007 | 28.603 | 0.000 | 0.181 | 0.207 |
Interpreting the model parameters#
Log-counts#
pr2.params["age"]
0.1938278482145407
Incidence rate ratio (IRR)#
np.exp(pr2.params["age"])
1.213887292102993
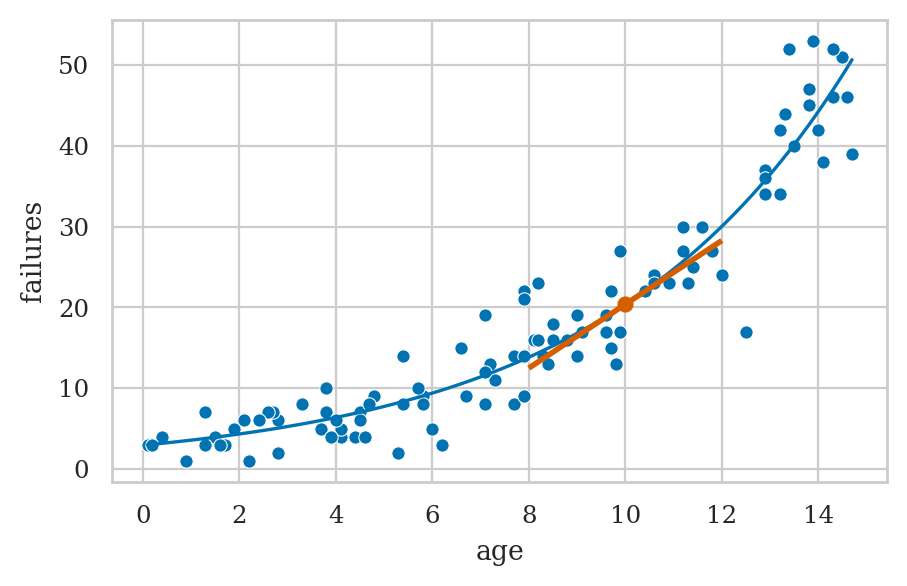
Marginal effect#
What is the marginal effect of the predictor age
for a 10 year old hard disk installation?
# using `statsmodels` .get_margeff() method
pr2.get_margeff(atexog={1:10}).summary_frame()
| dy/dx | Std. Err. | z | Pr(>|z|) | Conf. Int. Low | Cont. Int. Hi. | |
|---|---|---|---|---|---|---|
| age | 3.94912 | 0.151882 | 26.001165 | 4.804151e-149 | 3.651435 | 4.246804 |
# # ALT. manual calculation of the slope by evaluating the derivative
# b_0 = pr2.params['Intercept']
# b_age = pr2.params['age']
# np.exp(b_0 + b_age*10)*b_age
ax = plot_reg(pr2)
# Show the tangent line at age=10 (the marginal effect)
ptx, pty = (10, pr2.predict({"age":10})[0])
slope = pr2.get_margeff(atexog={1:10}).margeff[0]
dx = 2
endpoint1 = [ptx - dx, pty - dx*slope]
endpoint2 = [ptx + dx, pty + dx*slope]
lines = [[endpoint1, endpoint2]]
from matplotlib import collections as mc
lc = mc.LineCollection(lines, color="r", lw=2)
ax.plot([ptx], [pty], marker="o", color="r", alpha=1)
ax.add_collection(lc);
Predictions#
lam10 = pr2.predict({"age":10})[0]
lam10
20.374365915173986
from scipy.stats import poisson
Hhat = poisson(mu=lam10)
Hhat.ppf(0.05), Hhat.ppf(0.95)
(13.0, 28.0)
Explanations#
The exponential family of distributions#
exponential
Gaussian (normal)
Poisson
Binomial
The generalized linear model template#
choose
Generalized linear models using statsmodels#
import statsmodels.api as sm
Norm = sm.families.Gaussian()
Bin = sm.families.Binomial()
Pois = sm.families.Poisson()
Linear model#
students = pd.read_csv('datasets/students.csv')
formula0 = "score ~ 1 + effort"
glm0 = smf.glm(formula0, data=students, family=Norm).fit()
glm0.params
Intercept 32.465809
effort 4.504850
dtype: float64
Logistic regression#
formula1 = "hired ~ 1 + work"
glm1 = smf.glm(formula1, data=interns, family=Bin).fit()
glm1.params
# glm1.summary()
Intercept -78.693205
work 1.981458
dtype: float64
Poisson regression#
formula2 = "failures ~ 1 + age"
glm2 = smf.glm(formula2, data=hdisks, family=Pois).fit()
glm2.params
# glm2.summary()
Intercept 1.075999
age 0.193828
dtype: float64
Fitting generalized linear models#
Discussion#
Model diagnostics and validation#
# Dispersion from GLM attributes
# glm2.pearson_chi2 / glm2.df_resid
# Calculate Pearson chi-squared statistic
observed = hdisks['failures']
predicted = pr2.predict()
pearson_residuals = (observed - predicted) / np.sqrt(predicted)
pearson_chi2 = np.sum(pearson_residuals**2)
df_resid = pr2.df_resid
dispersion = pearson_chi2 / df_resid
print(f'Dispersion: {dispersion}')
# If dispersion > 1, consider Negative Binomial regression
Dispersion: 0.9869289289681199
Logistic regression as a building blocks for neural networks#
The operation of the perceptron, which is the basic building block of neural networks, is essentially the same as linear regression model:
constant intercept (bias term)
linear combination of inputs
nonlinear function used to force the output to be between 0 and 1
Limitations of GLMs#
GLMs assume observations are independent
Assumes distribution \(\mathcal{M}\) is one of the exponential family
Outliers can be problematic
Interpretability
Exercises#
Exercise 1: probabilities to odds and log-odds#
0.3/(1-0.3), 0.99/(1-0.99), 0.7/(1-0.7)
(0.4285714285714286, 98.99999999999991, 2.333333333333333)
logit(0.3), logit(0.99), logit(0.7)
(-0.8472978603872037, 4.595119850134589, 0.8472978603872034)
Exercise 2: log-odds to probabilities#
expit(-1), expit(1), expit(2)
(0.2689414213699951, 0.7310585786300049, 0.8807970779778823)
Exercise 3: students pass or fail#
a) Load the dataset students.csv and add a column passing
that contains 1 or 0, based on the above threshold score of 70.
students = pd.read_csv('datasets/students.csv')
students["passing"] = (students["score"] > 70).astype(int)
# students.head()
b) Fit a logistic regression model for passing based on effort variable.
lmpass = smf.logit("passing ~ 1 + effort", data=students).fit()
lmpass.params
Optimization terminated successfully.
Current function value: 0.276583
Iterations 8
Intercept -16.257302
effort 2.047882
dtype: float64
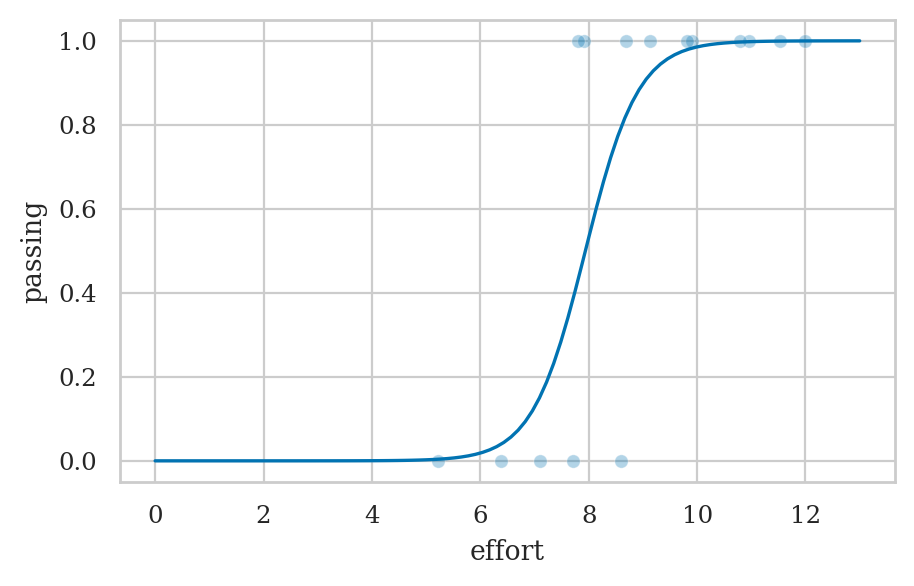
efforts = np.linspace(0, 13, 100)
passing_preds = lmpass.predict({"effort": efforts})
ax = sns.scatterplot(data=students, x="effort", y="passing", alpha=0.3)
sns.lineplot(x=efforts, y=passing_preds, ax=ax);
c) Use your model to predict the probability of a new student with effort 10 will pass.
lmpass.predict({"effort":10})
0 0.985536
dtype: float64
# ALT. compute prediction manually
intercept, b_effort = lmpass.params
expit(intercept + b_effort*10)
0.9855358765361845
Exercise 4: titanic survival data#
Fit a logistic regression model that calculates the probability of survival for people who were on the Titanic,
based on the data in datasets/exercises/titanic.csv. Use the variables age, sex, and pclass as predictors.
cf. Titanic_Logistic_Regression.ipynb
titanic = pd.read_csv('datasets/exercises/titanic.csv')
formula = "survived ~ age + C(sex) + C(pclass)"
lrtitanic = smf.logit(formula, data=titanic).fit()
lrtitanic.params
Optimization terminated successfully.
Current function value: 0.453279
Iterations 6
Intercept 3.777013
C(sex)[T.M] -2.522781
C(pclass)[T.2] -1.309799
C(pclass)[T.3] -2.580625
age -0.036985
dtype: float64
Use your logistic regression model to estimate the probability of survival for a 30 year old female traveling in second class.
pass30Fpclass2 = {"age":30, "sex":"F", "pclass":2}
lrtitanic.predict(pass30Fpclass2)
0 0.795378
dtype: float64
# # Cross check with sklearn
# from sklearn.linear_model import LogisticRegression
# df = pd.get_dummies(titanic, columns=['sex', 'pclass'], drop_first=True)
# X, y = df.drop('survived', axis=1), df['survived']
# sktitanic = LogisticRegression(penalty=None)
# sktitanic.fit(X, y)
# sktitanic.intercept_, sktitanic.coef_
Exercise 5: asthma attacks#
Fit a Poisson regression model to the datasets/exercises/asthma.csv dataset.
data source drkamarul/multivar_data_analysis
asthma = pd.read_csv("datasets/exercises/asthma.csv")
asthma
| gender | res_inf | ghq12 | attack | |
|---|---|---|---|---|
| 0 | female | yes | 21 | 6 |
| 1 | male | no | 17 | 4 |
| 2 | male | yes | 30 | 8 |
| 3 | female | yes | 22 | 5 |
| 4 | male | yes | 27 | 2 |
| ... | ... | ... | ... | ... |
| 115 | male | yes | 0 | 2 |
| 116 | female | yes | 31 | 2 |
| 117 | female | yes | 18 | 2 |
| 118 | female | yes | 21 | 3 |
| 119 | female | yes | 11 | 2 |
120 rows × 4 columns
formula_asthma = "attack ~ 1 + C(gender) + C(res_inf) + ghq12"
prasthma = smf.poisson(formula_asthma, data=asthma).fit()
prasthma.params
Optimization terminated successfully.
Current function value: 1.707281
Iterations 6
Intercept -0.315387
C(gender)[T.male] -0.041905
C(res_inf)[T.yes] 0.426431
ghq12 0.049508
dtype: float64
cf. https://bookdown.org/drki_musa/dataanalysis/poisson-regression.html#multivariable-analysis-1
Exercise 6: student admissions dataset#
The dataset datasets/exercises/binary.csv contains information
about the acceptance decision for 400 students to a prestigious school.
Try to fit a logistic regression model for the variable admit
using the variables gre, gpa, and rank as predictors.
# ORIGINAL https://stats.idre.ucla.edu/stat/data/binary.csv
binary = pd.read_csv('datasets/exercises/binary.csv')
binary.head(3)
| admit | gre | gpa | rank | |
|---|---|---|---|---|
| 0 | 0 | 380 | 3.61 | 3 |
| 1 | 1 | 660 | 3.67 | 3 |
| 2 | 1 | 800 | 4.00 | 1 |
lrbinary = smf.logit('admit ~ gre + gpa + C(rank)', data=binary).fit()
lrbinary.params
Optimization terminated successfully.
Current function value: 0.573147
Iterations 6
Intercept -3.989979
C(rank)[T.2] -0.675443
C(rank)[T.3] -1.340204
C(rank)[T.4] -1.551464
gre 0.002264
gpa 0.804038
dtype: float64
The above model uses the rank=1 as the reference category an the log odds reported are with respect to this catrgory
etc. for others rank[T.3] -1.340204 rank[T.4] -1.551464
See LogisticRegressionChangeOfReferenceCategoricalValue.ipynb for exercise recodign relative to different refrence level.
# # Cross check with sklearn
# from sklearn.linear_model import LogisticRegression
# df = pd.get_dummies(binary, columns=['rank'], drop_first=True)
# X, y = df.drop("admit", axis=1), df["admit"]
# lr = LogisticRegression(solver="lbfgs", penalty=None, max_iter=1000)
# lr.fit(X, y)
# lr.intercept_, lr.coef_
Exercise 7: ship accidents#
https://rdrr.io/cran/AER/man/ShipAccidents.html
https://pages.stern.nyu.edu/~wgreene/Text/tables/tablelist5.htm
https://pages.stern.nyu.edu/~wgreene/Text/tables/TableF21-3.txt
# TODO
Links#
TODO
CUT MATERIAL#
Bonus Exercises#
Bonus exercise A: honors class#
honors = pd.read_csv("datasets/exercises/honors.csv")
honors.sample(4)
| female | read | write | math | hon | femalexmath | |
|---|---|---|---|---|---|---|
| 172 | 1 | 42 | 36 | 42 | 0 | 42 |
| 157 | 1 | 42 | 57 | 51 | 0 | 51 |
| 75 | 0 | 47 | 40 | 43 | 0 | 0 |
| 178 | 1 | 50 | 52 | 45 | 0 | 45 |
Constant model#
lrhon1 = smf.logit("hon ~ 1", data=honors).fit()
lrhon1.params
Optimization terminated successfully.
Current function value: 0.556775
Iterations 5
Intercept -1.12546
dtype: float64
expit(lrhon1.params["Intercept"])
0.24499999999999994
honors["hon"].value_counts(normalize=True)
hon
0 0.755
1 0.245
Name: proportion, dtype: float64
Using only a categorical variable#
lrhon2 = smf.logit("hon ~ 1 + female", data=honors).fit()
lrhon2.params
Optimization terminated successfully.
Current function value: 0.549016
Iterations 5
Intercept -1.470852
female 0.592782
dtype: float64
pd.crosstab(honors["hon"], honors["female"], margins=True)
| female | 0 | 1 | All |
|---|---|---|---|
| hon | |||
| 0 | 74 | 77 | 151 |
| 1 | 17 | 32 | 49 |
| All | 91 | 109 | 200 |
b0 = lrhon2.params["Intercept"]
b_female = lrhon2.params["female"]
# male prob
expit(b0), 17/91
(0.18681318681318684, 0.18681318681318682)
# male odds
np.exp(b0), 17/74 # = (17/91) / (74/91)
(0.2297297297297298, 0.22972972972972974)
# male log-odds
b0, np.log(17/74)
(-1.4708517491479534, -1.4708517491479536)
# female prob
expit(b0 + b_female), 32/109
(0.29357798165137616, 0.29357798165137616)
# female odds
np.exp(b0 + b_female), 32/77 # = (32/109) / (77/109)
(0.41558441558441556, 0.4155844155844156)
b0 + b_female, np.log(32/77)
(-0.8780695190539574, -0.8780695190539572)
# odds female relative to male
np.exp(b_female)
1.8090145148968673
Logistic regression with a single continuous predictor variable#
lrhon3 = smf.logit("hon ~ 1 + math", data=honors).fit()
lrhon3.params
Optimization terminated successfully.
Current function value: 0.417683
Iterations 7
Intercept -9.793942
math 0.156340
dtype: float64
So the model equation is
# Increase in log-odds between math=54 and math=55
p54 = lrhon3.predict({"math":[54]}).item()
p55 = lrhon3.predict({"math":[55]}).item()
logit(p55) - logit(p54), lrhon3.params["math"]
(0.15634035558592307, 0.15634035558592288)
We can say now that the coefficient for math is the difference in the log odds. In other words, for a one-unit increase in the math score, the expected change in log odds is .1563404.
# Increase (multiplicative) in odds for unit increase in math
np.exp(lrhon3.params["math"]), (p55/(1-p55)) / (p54/(1-p54))
(1.1692240873242836, 1.1692240873242838)
So we can say for a one-unit increase in math score, we expect to see about 17% increase in the odds of being in an honors class. This 17% of increase does not depend on the value that math is held at.
Logistic regression with multiple predictor variables and no interaction terms#
lrhon4 = smf.logit("hon ~ 1 + math + female + read", data=honors).fit()
lrhon4.params
Optimization terminated successfully.
Current function value: 0.390424
Iterations 7
Intercept -11.770246
math 0.122959
female 0.979948
read 0.059063
dtype: float64
Logistic regression with an interaction term of two predictor variables#
lrhon5 = smf.logit("hon ~ 1 + math + female + femalexmath", data=honors).fit()
lrhon5.params
Optimization terminated successfully.
Current function value: 0.399417
Iterations 7
Intercept -8.745841
math 0.129378
female -2.899863
femalexmath 0.066995
dtype: float64
# ALT. without using `femalexmath` column
# lrhon5 = smf.logit("hon ~ 1 + math + female + female*math", data=honors).fit()
# lrhon5.params
Bonus exercise: LA high schools (NOT A VERY GOOD FIT FOR POISSON MODEL)#
Dataset info: http://www.philender.com/courses/intro/assign/data.html
This dataset consists of data from computer exercises collected from two high school in the Los Angeles area.
http://www.philender.com/courses/intro/code.html
lahigh_raw = pd.read_stata("https://stats.idre.ucla.edu/stat/stata/notes/lahigh.dta")
lahigh = lahigh_raw.convert_dtypes()
lahigh["gender"] = lahigh["gender"].astype(object).replace({1:"F", 2:"M"})
lahigh["ethnic"] = lahigh["ethnic"].astype(object).replace({
1:"Native American",
2:"Asian",
3:"African-American",
4:"Hispanic",
5:"White",
6:"Filipino",
7:"Pacific Islander"})
lahigh["school"] = lahigh["school"].astype(object).replace({1:"Alpha", 2:"Beta"})
lahigh.head()
| id | gender | ethnic | school | mathpr | langpr | mathnce | langnce | biling | daysabs | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1001 | M | Hispanic | Alpha | 63 | 36 | 56.988831 | 42.450859 | 2 | 4 |
| 1 | 1002 | M | Hispanic | Alpha | 27 | 44 | 37.094158 | 46.820587 | 2 | 4 |
| 2 | 1003 | F | Hispanic | Alpha | 20 | 38 | 32.275455 | 43.566574 | 2 | 2 |
| 3 | 1004 | F | Hispanic | Alpha | 16 | 38 | 29.056717 | 43.566574 | 2 | 3 |
| 4 | 1005 | F | Hispanic | Alpha | 2 | 14 | 6.748048 | 27.248474 | 3 | 3 |
formula = "daysabs ~ 1 + mathnce + langnce + C(gender)"
prlahigh = smf.poisson(formula, data=lahigh).fit()
prlahigh.params
Optimization terminated successfully.
Current function value: 4.898642
Iterations 5
Intercept 2.687666
C(gender)[T.M] -0.400921
mathnce -0.003523
langnce -0.012152
dtype: float64
# IRR
np.exp(prlahigh.params[1:])
C(gender)[T.M] 0.669703
mathnce 0.996483
langnce 0.987921
dtype: float64
# CI for IRR F
np.exp(prlahigh.conf_int().loc["C(gender)[T.M]"])
0 0.609079
1 0.736361
Name: C(gender)[T.M], dtype: float64
# prlahigh.summary()
# prlahigh.aic, prlahigh.bic
Diagnostics#
via https://www.statsmodels.org/dev/examples/notebooks/generated/postestimation_poisson.html
prdiag = prlahigh.get_diagnostic()
# Plot observed versus predicted frequencies for entire sample
# prdiag.plot_probs();
# Other:
# ['plot_probs',
# 'probs_predicted',
# 'results',
# 'test_chisquare_prob',
# 'test_dispersion',
# 'test_poisson_zeroinflation',
# 'y_max']
# Code to get exactly the same numbers as in
# https://stats.oarc.ucla.edu/stata/output/poisson-regression/
formula2 = "daysabs ~ 1 + mathnce + langnce + C(gender, Treatment(1))"
prlahigh2 = smf.poisson(formula2, data=lahigh).fit()
prlahigh2.params
Optimization terminated successfully.
Current function value: 4.898642
Iterations 5
Intercept 2.286745
C(gender, Treatment(1))[T.F] 0.400921
mathnce -0.003523
langnce -0.012152
dtype: float64