Section 5.4 — Bayesian difference between means#
This notebook contains the code examples from Section 5.4 Bayesian difference between means from the No Bullshit Guide to Statistics.
See also:
compare_iqs2_many_ways.ipynb
Examples: treszkai/best
Notebook setup#
# Ensure required Python modules are installed
%pip install --quiet numpy scipy seaborn statsmodels bambi==0.15.0 pymc==5.23.0 ministats
Note: you may need to restart the kernel to use updated packages.
# Load Python modules
import os
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import bambi as bmb
import arviz as az
# Figures setup
plt.clf() # needed otherwise `sns.set_theme` doesn't work
sns.set_theme(
context="paper",
style="whitegrid",
palette="colorblind",
rc={"font.family": "serif",
"font.serif": ["Palatino", "DejaVu Serif", "serif"],
"figure.figsize": (5,3)},
)
%config InlineBackend.figure_format = "retina"
<Figure size 640x480 with 0 Axes>
# Simple float __repr__
if int(np.__version__.split(".")[0]) >= 2:
np.set_printoptions(legacy='1.25')
# Download datasets/ directory if necessary
from ministats import ensure_datasets
ensure_datasets()
datasets/ directory already exists.
Introduciton#
We’ll now revisit the problem of comparison between two groups from a Bayesian perspective.
Model#
The data models for the two groups are based on Student’s \(t\)-distribution:
We’ll use a normal prior for the means, half-\(t\) priors for standard deviations, and a gamma distribution for the degrees of freedom parameter:
Bambi formula objects#
import bambi as bmb
formula = bmb.Formula("y ~ 0 + group",
"sigma ~ 0 + group")
formula
Formula('y ~ 0 + group', 'sigma ~ 0 + group')
formula.main, formula.additionals
('y ~ 0 + group', ('sigma ~ 0 + group',))
formula.get_all_formulas()
['y ~ 0 + group', 'sigma ~ 0 + group']
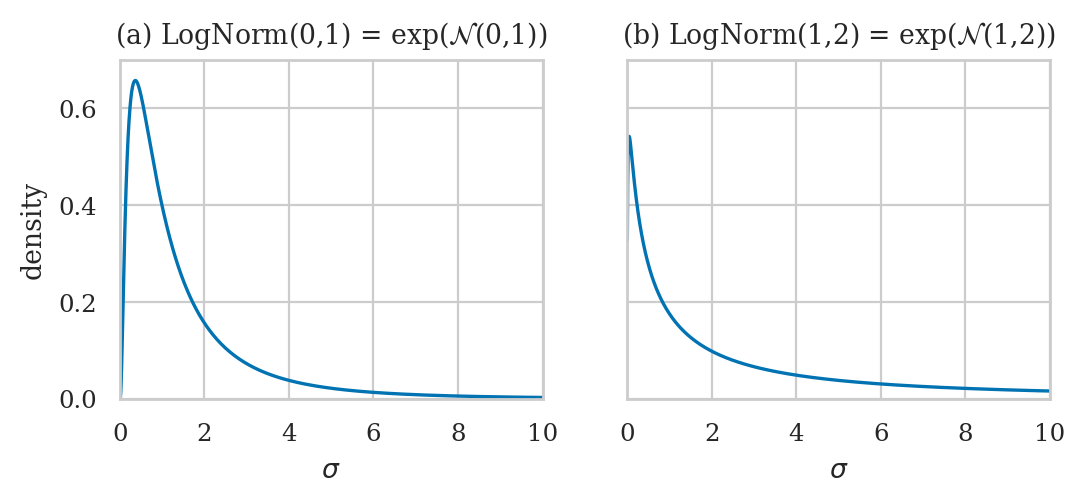
Choosing priors for log-sigma#
Normal distribution for log-sigma#
from scipy.stats import norm
def get_lognormal(mu=0, sigma=1):
logsigs = np.linspace(-5, 5, 1000)
dlogsigs = norm(mu, sigma).pdf(logsigs)
sigmas = np.exp(logsigs)
# Apply the change of variables to get the density for sigma
# based on the Jacobian |d(log(sigma))/d(sigma)| = 1/sigma
dsigmas = dlogsigs / sigmas
return sigmas, dsigmas
with plt.rc_context({"figure.figsize":(6,2.2)}):
fig, (ax1, ax2) = plt.subplots(1,2, sharey=True)
# PLOT 1
mu, sigma = 0, 1
sigmas, dsigmas = get_lognormal(mu, sigma)
sns.lineplot(x=sigmas, y=dsigmas, ax=ax1)
ax1.set_xlabel('$\\sigma$')
ax1.set_ylabel('density')
ax1.set_title(f'(a) LogNorm({mu},{sigma}) = exp($\\mathcal{{N}}$({mu},{sigma}))')
ax1.set_xlim(0, 10)
ax1.set_ylim(0, 0.7)
# PLOT 2
mu, sigma = 1, 2
sigmas, dsigmas = get_lognormal(mu, sigma)
sns.lineplot(x=sigmas, y=dsigmas, ax=ax2)
ax2.set_xlabel('$\\sigma$')
ax2.set_ylabel('density')
ax2.set_title(f'(b) LogNorm({mu},{sigma}) = exp($\\mathcal{{N}}$({mu},{sigma}))')
ax2.set_xlim(0, 10)
Choosing priors for the degrees of freedom parameter#
# FIGURES ONLY
from scipy.stats import gamma as gammadist
from scipy.stats import expon
from ministats import plot_pdf
with plt.rc_context({"figure.figsize":(6,2.2)}):
fig, (ax1, ax2) = plt.subplots(1,2, sharey=True)
# PLOT 1
alpha, beta = 2, 0.1
rv_nu = gammadist(a=2, scale=1/beta)
plot_pdf(rv_nu, rv_name="ν", ax=ax1)
ax1.set_title(f'(a) Gamma($\\alpha=${alpha}, $\\beta=${beta})')
ax1.set_xlim(0, 100)
# PLOT 2
scale = 30
rv_nu_exp = expon(scale=scale)
plot_pdf(rv_nu_exp, rv_name="ν", ax=ax2)
ax2.set_title(f'(b) Expon($\\lambda=$1/{scale})')
ax2.set_xlim(0, 100)
Example 1: comparing electricity prices#
Electricity prices from East End and West End
Electricity prices dataset#
eprices = pd.read_csv("datasets/eprices.csv")
eprices.groupby("loc")["price"].describe()
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| loc | ||||||||
| East | 9.0 | 6.155556 | 0.877655 | 4.8 | 5.5 | 6.3 | 6.5 | 7.7 |
| West | 9.0 | 9.155556 | 1.562139 | 6.8 | 8.3 | 8.6 | 10.0 | 11.8 |
eprices["price"].mean(), eprices["price"].std()
(7.655555555555556, 1.973120020267162)
Bayesian model#
Bambi model#
formula1 = bmb.Formula("price ~ 0 + loc",
"sigma ~ 0 + loc")
links1 = {"mu":"identity",
"sigma":"log"}
priors1 = {
"loc": bmb.Prior("Normal", mu=8, sigma=5),
"sigma": {
"loc": bmb.Prior("Normal", mu=0, sigma=1)
},
"nu": bmb.Prior("Gamma", alpha=2, beta=0.1),
}
mod1 = bmb.Model(formula=formula1,
family="t",
link=links1,
priors=priors1,
data=eprices)
mod1
Formula: price ~ 0 + loc
sigma ~ 0 + loc
Family: t
Link: mu = identity
sigma = log
Observations: 18
Priors:
target = mu
Common-level effects
loc ~ Normal(mu: 8.0, sigma: 5.0)
Auxiliary parameters
nu ~ Gamma(alpha: 2.0, beta: 0.1)
target = sigma
Common-level effects
sigma_loc ~ Normal(mu: 0.0, sigma: 1.0)
mod1.build()
mod1.graph()
Prior predictive checks#
# TODO
Model fitting and analysis#
idata1 = mod1.fit(random_seed=42)
Initializing NUTS using jitter+adapt_diag...
/opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/site-packages/pytensor/link/c/cmodule.py:2968: UserWarning: PyTensor could not link to a BLAS installation. Operations that might benefit from BLAS will be severely degraded.
This usually happens when PyTensor is installed via pip. We recommend it be installed via conda/mamba/pixi instead.
Alternatively, you can use an experimental backend such as Numba or JAX that perform their own BLAS optimizations, by setting `pytensor.config.mode == 'NUMBA'` or passing `mode='NUMBA'` when compiling a PyTensor function.
For more options and details see https://pytensor.readthedocs.io/en/latest/troubleshooting.html#how-do-i-configure-test-my-blas-library
warnings.warn(
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [nu, loc, sigma_loc]
Sampling 2 chains for 1_000 tune and 1_000 draw iterations (2_000 + 2_000 draws total) took 2 seconds.
We recommend running at least 4 chains for robust computation of convergence diagnostics
Processing the results#
Calculate derived quantities used for the analysis plots and summaries.
post1 = idata1["posterior"]
# Calculate sigmas from log-sigmas
#######################################################
logsig_W = post1["sigma_loc"].sel(sigma_loc_dim="West")
logsig_E = post1["sigma_loc"].sel(sigma_loc_dim="East")
post1["sigma_West"] = np.exp(logsig_W)
post1["sigma_East"] = np.exp(logsig_E)
# Calculate the difference between between means
post1["mu_West"] = post1["loc"].sel(loc_dim="West")
post1["mu_East"] = post1["loc"].sel(loc_dim="East")
post1["dmeans"] = post1["mu_West"] - post1["mu_East"]
# Calculate the difference between standard deviations
post1["dsigmas"] = post1["sigma_West"]-post1["sigma_East"]
# Effect size
#######################################################
pvar =(post1["sigma_West"]**2+post1["sigma_East"]**2)/2
post1["cohend"] = post1["dmeans"] / np.sqrt(pvar)
# #ALT: use helper function
# from ministats import calc_dmeans_stats
# calc_dmeans_stats(idata1, group_name="loc")
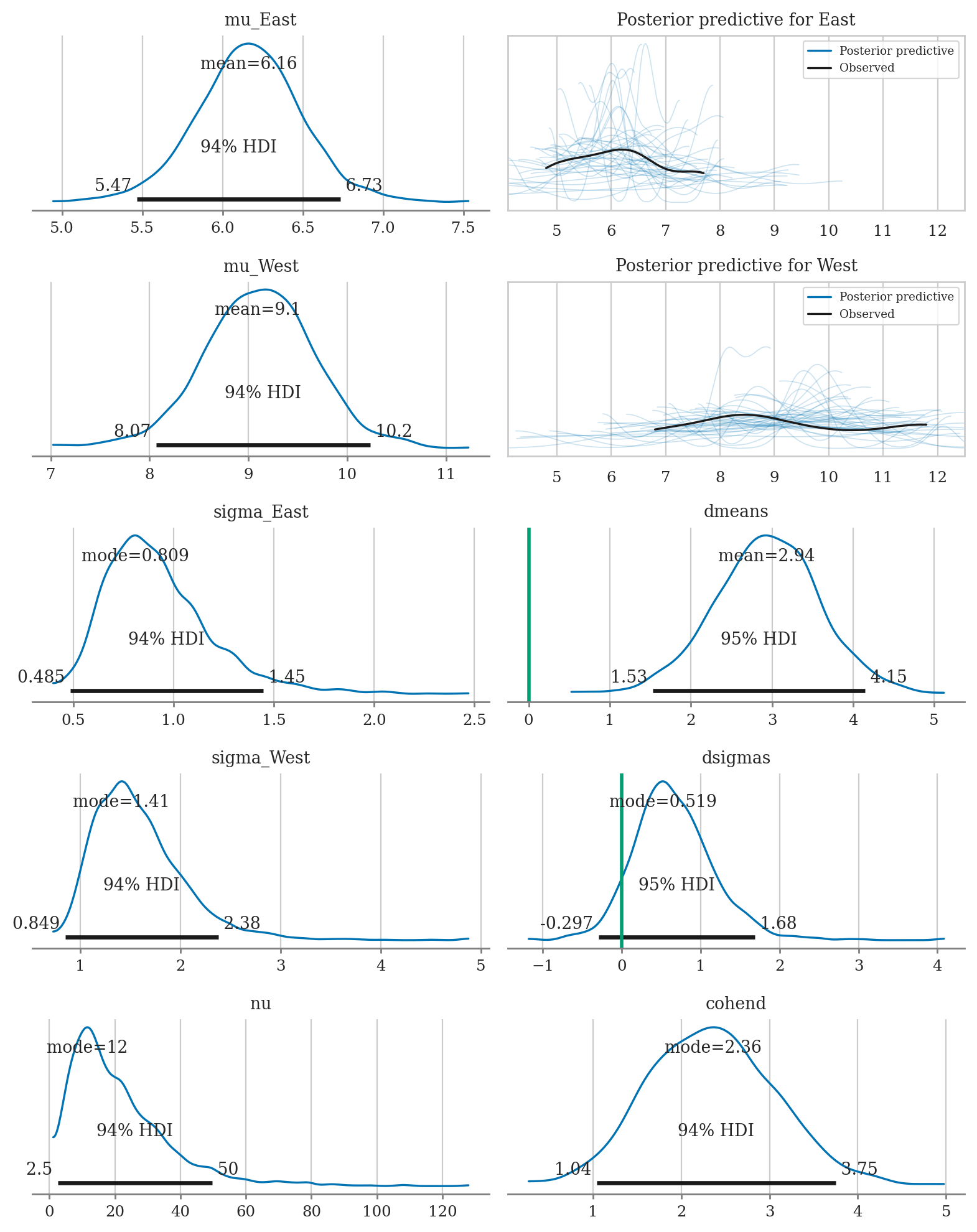
az.summary(idata1, kind="stats", hdi_prob=0.90,
var_names=["mu_West", "mu_East", "dmeans", "sigma_West", "sigma_East", "dsigmas", "nu", "cohend"])
| mean | sd | hdi_5% | hdi_95% | |
|---|---|---|---|---|
| mu_West | 9.099 | 0.571 | 8.221 | 10.041 |
| mu_East | 6.163 | 0.333 | 5.616 | 6.681 |
| dmeans | 2.936 | 0.660 | 1.853 | 4.027 |
| sigma_West | 1.583 | 0.459 | 0.929 | 2.223 |
| sigma_East | 0.927 | 0.276 | 0.546 | 1.342 |
| dsigmas | 0.656 | 0.520 | -0.183 | 1.440 |
| nu | 21.451 | 15.481 | 2.189 | 41.225 |
| cohend | 2.351 | 0.726 | 1.276 | 3.611 |
Compare to frequentist results#
from scipy.stats import ttest_ind
pricesW = eprices[eprices["loc"]=="West"]["price"]
pricesE = eprices[eprices["loc"]=="East"]["price"]
res1 = ttest_ind(pricesW, pricesE, equal_var=False)
res1.statistic, res1.pvalue
(5.022875513276464, 0.0002570338337217609)
res1.confidence_interval(confidence_level=0.9)
ConfidenceInterval(low=1.9396575883681466, high=4.060342411631854)
from ministats import cohend2
cohend2(pricesW, pricesE)
2.3678062243290983
Conclusions#
Example 2: comparing IQ scores#
We’ll look at IQ scores data taken from a the paper Bayesian Estimation Supersedes the t-Test (BEST) by John K. Kruschke.
smart drug administered to treatment group and want to compare to control group. Data contains outliers)
cf. compare_iqs2_many_ways.ipynb
IQ scores dataset#
iqs2 = pd.read_csv("datasets/iqs2.csv")
iqs2.groupby("group")["iq"].describe()
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| group | ||||||||
| ctrl | 42.0 | 100.357143 | 2.516496 | 88.0 | 100.0 | 100.5 | 101.0 | 105.0 |
| treat | 47.0 | 101.914894 | 6.021085 | 82.0 | 100.0 | 102.0 | 103.0 | 124.0 |
Bayesian model#
TODO: add formulas
Bambi model#
formula2 = bmb.Formula("iq ~ 0 + group",
"sigma ~ 0 + group")
priors2 = {
"group": bmb.Prior("Normal", mu=100, sigma=35),
"sigma": {
"group": bmb.Prior("Normal", mu=1, sigma=2)
},
"nu": bmb.Prior("Gamma", alpha=2, beta=0.1),
}
mod2 = bmb.Model(formula=formula2,
family="t",
# link={"mu":"identity", "sigma":"log"}, # Bambi defaults
priors=priors2,
data=iqs2)
mod2
Formula: iq ~ 0 + group
sigma ~ 0 + group
Family: t
Link: mu = identity
sigma = log
Observations: 89
Priors:
target = mu
Common-level effects
group ~ Normal(mu: 100.0, sigma: 35.0)
Auxiliary parameters
nu ~ Gamma(alpha: 2.0, beta: 0.1)
target = sigma
Common-level effects
sigma_group ~ Normal(mu: 1.0, sigma: 2.0)
mod2.build()
mod2.graph()
Model fitting and analysis#
idata2 = mod2.fit(random_seed=42)
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [nu, group, sigma_group]
Sampling 2 chains for 1_000 tune and 1_000 draw iterations (2_000 + 2_000 draws total) took 2 seconds.
We recommend running at least 4 chains for robust computation of convergence diagnostics
from ministats import calc_dmeans_stats
calc_dmeans_stats(idata2, group_name="group");
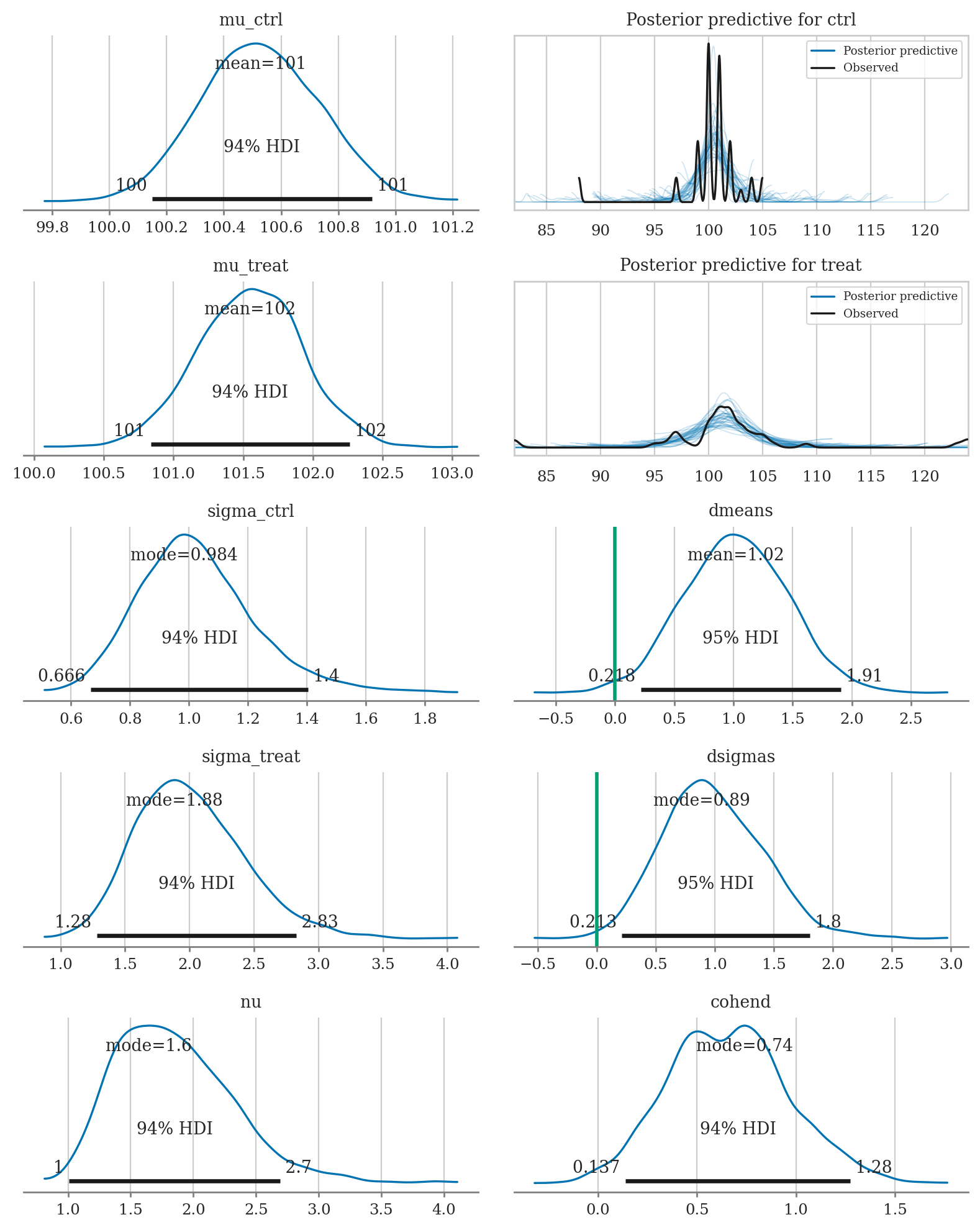
az.summary(idata2, kind="stats", hdi_prob=0.95,
var_names=["mu_treat", "mu_ctrl", "dmeans", "sigma_treat", "sigma_ctrl", "dsigmas", "nu", "cohend"])
| mean | sd | hdi_2.5% | hdi_97.5% | |
|---|---|---|---|---|
| mu_treat | 101.547 | 0.379 | 100.807 | 102.285 |
| mu_ctrl | 100.528 | 0.209 | 100.150 | 100.947 |
| dmeans | 1.019 | 0.441 | 0.218 | 1.907 |
| sigma_treat | 2.006 | 0.422 | 1.228 | 2.839 |
| sigma_ctrl | 1.026 | 0.201 | 0.663 | 1.437 |
| dsigmas | 0.980 | 0.426 | 0.213 | 1.803 |
| nu | 1.859 | 0.487 | 1.002 | 2.773 |
| cohend | 0.660 | 0.307 | 0.091 | 1.291 |
Sensitivity analysis#
from ministats.book.tables import sens_analysis_dmeans_iqs2
results = sens_analysis_dmeans_iqs2(iqs2)
results
| M_prior | logSigma_prior | Nu_prior | dmeans_mean | dmeans_95hdi | dsigmas_mode | dsigmas_95hdi | nu_mode | codhend_mode | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | $\mathcal{N}(100,35)$ | $\mathcal{N}(1,2)$ | $\Gamma(2,0.1)$ | 1.02 | [0.232, 1.907] | 0.897 | [0.213, 1.858] | 1.672 | 0.897 |
| 1 | $\mathcal{N}(100,50)$ | $\mathcal{N}(1,2)$ | $\Gamma(2,0.1)$ | 1.014 | [0.207, 1.888] | 0.932 | [0.143, 1.823] | 1.71 | 0.932 |
| 2 | $\mathcal{N}(100,10)$ | $\mathcal{N}(1,2)$ | $\Gamma(2,0.1)$ | 1.026 | [0.227, 1.917] | 0.905 | [0.201, 1.829] | 1.724 | 0.905 |
| 3 | $\mathcal{N}(100,35)$ | $\mathcal{N}(0,1)$ | $\Gamma(2,0.1)$ | 1.027 | [0.193, 1.838] | 0.831 | [0.178, 1.714] | 1.608 | 0.831 |
| 4 | $\mathcal{N}(100,35)$ | $\mathcal{N}(1,2)$ | $\textrm{Expon}(1/30)$ | 1.027 | [0.261, 1.867] | 0.795 | [0.217, 1.882] | 1.6 | 0.795 |
Compare to frequentist results#
from scipy.stats import ttest_ind
treated = iqs2[iqs2["group"]=="treat"]["iq"]
controls = iqs2[iqs2["group"]=="ctrl"]["iq"]
res2 = ttest_ind(treated, controls, equal_var=False)
res2.statistic, res2.pvalue
(1.622190457290228, 0.10975381983712831)
res2.confidence_interval(confidence_level=0.95)
ConfidenceInterval(low=-0.36118477236636837, high=3.4766862921232016)
from ministats import cohend2
cohend2(treated, controls)
0.3309654530139732
# MAYBE
# # Test if the variances of the two groups are the same
# from scipy.stats import levene
# levene(treated, controls)
Conclusions#
Explanations#
Alternative choices of priors#
e.g. prior for sigma as very wiiiiiide, and nu ~ Exp(1/29)+1 → BEST Bayesian estimation supersedes the t-test by John K. Kruschke
If we use a model with common standard deviation, we get equivalent of pooled sigma
# TODO
Performance tests#
Simulated datasets#
Simulate datasets with various combinations of \(n\), \(\Delta\), and outliers.
from ministats.book.tables import gen_dmeans_datasets
ns = [20, 30, 50, 100]
Deltas = [0, 0.2, 0.5, 0.8, 1.3]
outliers_options = ["no", "few", "lots"]
dataset_specs = gen_dmeans_datasets(ns=ns,
Deltas=Deltas,
outliers_options=outliers_options)
dataset_specs[0:4]
[{'n': 20, 'Delta': 0, 'outliers': 'no', 'random_seed': 45},
{'n': 20, 'Delta': 0, 'outliers': 'few', 'random_seed': 46},
{'n': 20, 'Delta': 0, 'outliers': 'lots', 'random_seed': 47},
{'n': 20, 'Delta': 0.2, 'outliers': 'no', 'random_seed': 48}]
Model comparisons#
Compare results several models:
Classical permutation test (from Section 4.5)
Classical Welch’s two-sample t-test (from Section 4.5)
Bayesian model with \(\mathcal{N}\) data model
Robust Bayesian model with \(\mathcal{T}\) data model
Bayes factors with JZS prior and standard cutoff
from ministats.book.tables import fit_dmeans_models
help(fit_dmeans_models)
Help on function fit_dmeans_models in module ministats.book.tables:
fit_dmeans_models(dataset, random_seed=42)
Fit the following models for the difference of two means:
- Permutation test from Section 3.5
- Welch's two-sample $t$-test from Section 3.5
- Bayesian model that uses normal as data model
- Robust Bayesian model that uses t-distribution as data model
- Bayes factor using JZS prior (using `pingouin` library)
For each model, we run the hypothesis test to decide if the populations
are the same or different based on the conventional cutoff level of 5%.
We also construct a 90% interval estimates for the unknown `Delta`.
Results#
# Download simdata/ directory if necessary
from ministats import ensure_simdata
ensure_simdata()
# Option A: load results from simulation
filename = "dmeans_perf_metrics__" \
+ "ns_20_30_50_100__" \
+ "Deltas_0_0.2_0.5_0.8_1.3__" \
+ "outs_no_few_lots__reps_100.csv"
results = pd.read_csv(os.path.join("simdata", filename), index_col=[0,1])
results.head(3)
# Option B: run full simulation (takes about three days)
# from ministats.book.tables import calc_dmeans_perf_metrics
# results = calc_dmeans_perf_metrics(reps=100)
simdata/ directory present and ready.
| n | Delta | outliers | seed | count_reject | count_fail_to_reject | count_captured | avg_width | ||
|---|---|---|---|---|---|---|---|---|---|
| spec | model | ||||||||
| 0 | perm | 20 | 0.0 | no | 45 | 6 | 94 | 90.0 | 1.011714 |
| welch | 20 | 0.0 | no | 45 | 6 | 94 | 90.0 | 1.065525 | |
| norm_bayes | 20 | 0.0 | no | 45 | 2 | 98 | 94.0 | 1.294730 |
Type I (false positive) error rates#
from ministats.book.tables import get_perf_table_typeI
tableA = get_perf_table_typeI(results)
tableA
| model | perm | welch | norm_bayes | robust_bayes | bf | |
|---|---|---|---|---|---|---|
| outliers | n | |||||
| no | 20 | 0.06 | 0.06 | 0.02 | 0.07 | 0.02 |
| 30 | 0.04 | 0.04 | 0.03 | 0.04 | 0.03 | |
| 50 | 0.04 | 0.04 | 0.01 | 0.04 | 0.01 | |
| 100 | 0.06 | 0.06 | 0.03 | 0.05 | 0.01 | |
| few | 20 | 0.04 | 0.04 | 0.01 | 0.05 | 0.01 |
| 30 | 0.04 | 0.04 | 0.01 | 0.05 | 0.00 | |
| 50 | 0.03 | 0.03 | 0.02 | 0.01 | 0.00 | |
| 100 | 0.07 | 0.07 | 0.06 | 0.08 | 0.03 | |
| lots | 20 | 0.04 | 0.02 | 0.01 | 0.03 | 0.01 |
| 30 | 0.02 | 0.02 | 0.00 | 0.02 | 0.00 | |
| 50 | 0.07 | 0.06 | 0.06 | 0.07 | 0.04 | |
| 100 | 0.03 | 0.03 | 0.03 | 0.02 | 0.00 |
Observations:
all models are about the same
BF model has significantly less false positives
Power analysis#
from ministats.book.tables import get_perf_table_power
tableB = get_perf_table_power(results, show_all=False)
tableB
| model | perm | welch | norm_bayes | robust_bayes | bf | ||
|---|---|---|---|---|---|---|---|
| outliers | Delta | n | |||||
| no | 0.5 | 30 | 0.46 | 0.46 | 0.33 | 0.46 | 0.31 |
| 50 | 0.68 | 0.69 | 0.54 | 0.66 | 0.50 | ||
| 100 | 0.93 | 0.93 | 0.90 | 0.93 | 0.84 | ||
| 0.8 | 20 | 0.60 | 0.60 | 0.44 | 0.59 | 0.49 | |
| 30 | 0.86 | 0.86 | 0.77 | 0.86 | 0.72 | ||
| 50 | 0.99 | 0.99 | 0.96 | 0.99 | 0.93 | ||
| few | 0.5 | 30 | 0.49 | 0.49 | 0.32 | 0.45 | 0.29 |
| 50 | 0.49 | 0.51 | 0.39 | 0.58 | 0.33 | ||
| 100 | 0.80 | 0.80 | 0.80 | 0.88 | 0.69 | ||
| 0.8 | 20 | 0.69 | 0.68 | 0.51 | 0.65 | 0.54 | |
| 30 | 0.88 | 0.88 | 0.79 | 0.87 | 0.78 | ||
| 50 | 0.85 | 0.85 | 0.83 | 0.94 | 0.75 | ||
| lots | 0.5 | 30 | 0.33 | 0.32 | 0.29 | 0.43 | 0.20 |
| 50 | 0.52 | 0.52 | 0.50 | 0.63 | 0.33 | ||
| 100 | 0.64 | 0.63 | 0.60 | 0.88 | 0.52 | ||
| 0.8 | 20 | 0.44 | 0.43 | 0.37 | 0.63 | 0.30 | |
| 30 | 0.60 | 0.59 | 0.57 | 0.81 | 0.48 | ||
| 50 | 0.82 | 0.83 | 0.79 | 0.97 | 0.66 |
Observations:
Bayes \(\mathcal{T}\) has higher power than Bayes \(\mathcal{N}\)
Bayes \(\mathcal{T}\) outperforms all other models when outliers are present
Interval estimates coverage#
from ministats.book.tables import get_perf_table_coverage
tableC = get_perf_table_coverage(results)
tableC
| model | perm | welch | norm_bayes | robust_bayes | |||||
|---|---|---|---|---|---|---|---|---|---|
| coverage | avg_width | coverage | avg_width | coverage | avg_width | coverage | avg_width | ||
| outliers | n | ||||||||
| no | 20 | 0.8800 | 1.011711 | 0.9050 | 1.064862 | 0.9650 | 1.298070 | 0.9100 | 1.078473 |
| 30 | 0.9100 | 0.821001 | 0.9175 | 0.849298 | 0.9625 | 1.016220 | 0.9225 | 0.855460 | |
| 50 | 0.9050 | 0.651732 | 0.9100 | 0.665610 | 0.9525 | 0.762513 | 0.9100 | 0.667750 | |
| 100 | 0.9025 | 0.462228 | 0.9025 | 0.467091 | 0.9325 | 0.517568 | 0.9075 | 0.467747 | |
| few | 20 | 0.8550 | 1.002797 | 0.8675 | 1.056252 | 0.9500 | 1.294572 | 0.8700 | 1.069132 |
| 30 | 0.9175 | 0.835295 | 0.9250 | 0.864310 | 0.9625 | 1.022407 | 0.9250 | 0.872108 | |
| 50 | 0.8675 | 0.784709 | 0.9025 | 0.802027 | 0.9350 | 0.854340 | 0.9075 | 0.695260 | |
| 100 | 0.8725 | 0.554332 | 0.8975 | 0.560776 | 0.9125 | 0.581300 | 0.9275 | 0.487010 | |
| lots | 20 | 0.8700 | 1.442238 | 0.9025 | 1.530370 | 0.9450 | 1.601472 | 0.8975 | 1.197020 |
| 30 | 0.8550 | 1.085291 | 0.8700 | 1.128444 | 0.9000 | 1.197037 | 0.8550 | 0.926892 | |
| 50 | 0.8825 | 0.892523 | 0.8975 | 0.913113 | 0.9175 | 0.937645 | 0.9075 | 0.726270 | |
| 100 | 0.8975 | 0.679404 | 0.9025 | 0.686951 | 0.9175 | 0.688840 | 0.8950 | 0.509295 | |
Observations:
All methods have coverage close to the nominal 90%
Bayes \(\mathcal{T}\) produces narrower intervals
Discussion#
Comparison to the frequentist two-sample t-test#
results numerically similar
note we’re using 𝒯 as data model, not as a sampling distribution
conceptually different:
p-value vs. decision based on posterior distribution
confidence intervals vs. credible intervals
Comparing multiple groups#
?Extension to Bayesian ANOVA? Can extend approach to multiple groups: Bayesian ANOVA
FWD reference to hierarchical models for group comparison covered in Section 5.5
Exercises#
Exercise 1: small samples#
As = [5.77, 5.33, 4.59, 4.33, 3.66, 4.48]
Bs = [3.88, 3.55, 3.29, 2.59, 2.33, 3.59]
groups = ["A"]*len(As) + ["B"]*len(Bs)
df1 = pd.DataFrame({"group": groups, "vals": As + Bs})
# df1
Exercise 2: lecture and debate curriculums#
students = pd.read_csv("datasets/students.csv")
# students.groupby("curriculum")["score"].describe()
formula_std = bmb.Formula("score ~ 0 + curriculum",
"sigma ~ 0 + curriculum")
priors_std = {
"curriculum": bmb.Prior("Normal", mu=70, sigma=30),
"sigma": {
"curriculum":bmb.Prior("Normal", mu=1, sigma=2)
},
"nu": bmb.Prior("Gamma", alpha=2, beta=0.1),
}
mod_std = bmb.Model(formula=formula_std,
family="t",
link="identity",
priors=priors_std,
data=students)
mod_std
# mod_std.build()
# mod_std.graph()
Formula: score ~ 0 + curriculum
sigma ~ 0 + curriculum
Family: t
Link: mu = identity
sigma = log
Observations: 15
Priors:
target = mu
Common-level effects
curriculum ~ Normal(mu: 70.0, sigma: 30.0)
Auxiliary parameters
nu ~ Gamma(alpha: 2.0, beta: 0.1)
target = sigma
Common-level effects
sigma_curriculum ~ Normal(mu: 1.0, sigma: 2.0)
idata_std = mod_std.fit(random_seed=42)
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [nu, curriculum, sigma_curriculum]
Sampling 2 chains for 1_000 tune and 1_000 draw iterations (2_000 + 2_000 draws total) took 2 seconds.
We recommend running at least 4 chains for robust computation of convergence diagnostics
# Calculate the difference between between means
currs = idata_std["posterior"]["curriculum"]
mu_debate = currs.sel(curriculum_dim="debate")
mu_lecture = currs.sel(curriculum_dim="lecture")
idata_std["posterior"]["dmeans"] = mu_debate - mu_lecture
az.summary(idata_std, kind="stats", var_names=["dmeans"], hdi_prob=0.95)
| mean | sd | hdi_2.5% | hdi_97.5% | |
|---|---|---|---|---|
| dmeans | 7.685 | 5.428 | -3.012 | 18.201 |
# from ministats import plot_dmeans_stats
# plot_dmeans_stats(mod_std, idata_std, ppc_xlims=[50,100], group_name="curriculum");
# from scipy.stats import ttest_ind
# scoresD = students[students["curriculum"]=="debate"]["score"]
# scoresL = students[students["curriculum"]=="lecture"]["score"]
# res_std = ttest_ind(scoresD, scoresL, equal_var=False)
# res_std.statistic, res_std.pvalue
# from ministats import cohend2
# cohend2(scoresL, scoresD)
Exercise 3: redo exercises from Section 3.5 section using Bayesian methods#
# TODO
Links#
TODO
BONUS Examples#
Example 4: small example form BEST vignette#
See http://cran.nexr.com/web/packages/BEST/vignettes/BEST.pdf#page=2
# y1s = [5.77, 5.33, 4.59, 4.33, 3.66, 4.48]
# y2s = [3.88, 3.55, 3.29, 2.59, 2.33, 3.59]
# from ministats.bayes import bayes_dmeans
# mod4, idata4 = bayes_dmeans(y1s, y2s, groups=["y1", "y2"])
# from ministats import calc_dmeans_stats
# calc_dmeans_stats(idata4)
# az.summary(idata4, kind="stats", hdi_prob=0.95,
# var_names=["dmeans", "sigma_y1", "sigma_y2", "dsigmas", "nu", "cohend"])
# from ministats import plot_dmeans_stats
# plot_dmeans_stats(mod4, idata4, ppc_xlims=None);
Example 5: comparing morning to evening#
# morning = [8.99, 9.21, 9.03, 9.15, 8.68, 8.82, 8.66, 8.82, 8.59, 8.14,
# 9.09, 8.80, 8.18, 9.23, 8.55, 9.03, 9.36, 9.06, 9.57, 8.38]
# evening = [9.82, 9.34, 9.73, 9.93, 9.33, 9.41, 9.48, 9.14, 8.62, 8.60,
# 9.60, 9.41, 8.43, 9.77, 8.96, 9.81, 9.75, 9.50, 9.90, 9.13]
# from ministats.bayes import bayes_dmeans
# mod5, idata5 = bayes_dmeans(evening, morning, groups=["evening", "morning"])
# from ministats import calc_dmeans_stats
# calc_dmeans_stats(idata5)
# az.summary(idata5, kind="stats", hdi_prob=0.95,
# var_names=["dmeans", "sigma_evening", "sigma_morning", "dsigmas", "nu", "cohend"])
# from ministats import plot_dmeans_stats
# plot_dmeans_stats(mod5, idata5, ppc_xlims=None);