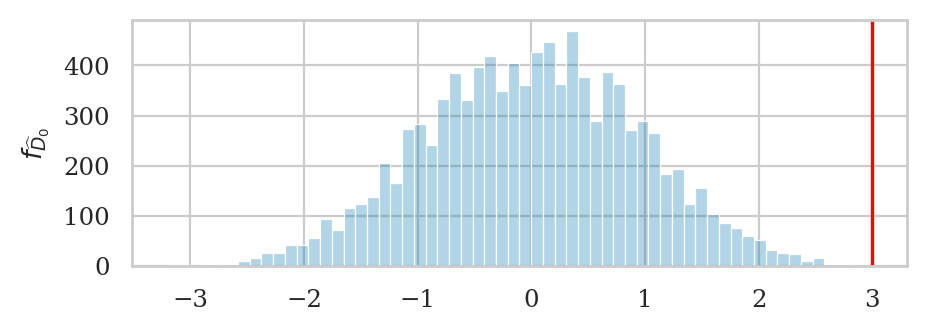Section 3.5 — Two-sample hypothesis tests#
This notebook contains the code examples from Section 3.5 Two-sample hypothesis tests of the No Bullshit Guide to Statistics.
Notebook setup#
# Ensure required Python modules are installed
%pip install --quiet numpy scipy seaborn pandas ministats
Note: you may need to restart the kernel to use updated packages.
# load Python modules
import os
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# Plot helper functions
from ministats import nicebins
from ministats import plot_pdf
from ministats.utils import savefigure
# Stats helper functions
from ministats import tailvalues
from ministats import tailprobs
# Figures setup
plt.clf() # needed otherwise `sns.set_theme` doesn't work
sns.set_theme(
context="paper",
style="whitegrid",
palette="colorblind",
rc={"font.family": "serif",
"font.serif": ["Palatino", "DejaVu Serif", "serif"],
"figure.figsize": (5, 1.6)},
)
%config InlineBackend.figure_format = 'retina'
<Figure size 640x480 with 0 Axes>
# Simple float __repr__
if int(np.__version__.split(".")[0]) >= 2:
np.set_printoptions(legacy='1.25')
# set random seed for repeatability
np.random.seed(42)
# Download datasets/ directory if necessary
from ministats import ensure_datasets
ensure_datasets()
datasets/ directory already exists.
\(\def\stderr#1{\mathbf{se}_{#1}}\) \(\def\stderrhat#1{\hat{\mathbf{se}}_{#1}}\) \(\newcommand{\Mean}{\textbf{Mean}}\) \(\newcommand{\Var}{\textbf{Var}}\) \(\newcommand{\Std}{\textbf{Std}}\) \(\newcommand{\Freq}{\textbf{Freq}}\) \(\newcommand{\RelFreq}{\textbf{RelFreq}}\) \(\newcommand{\DMeans}{\textbf{DMeans}}\) \(\newcommand{\Prop}{\textbf{Prop}}\) \(\newcommand{\DProps}{\textbf{DProps}}\)
(this cell contains the macro definitions like \(\stderr{\overline{\mathbf{x}}}\), \(\stderrhat{}\), \(\Mean\), …)
Definitions#
Populations. We’ll label the two unknown populations \(X\) and \(Y\). and assume the arbitrary model family \(\mathcal{M}\) with mean parameter \(X \sim \mathcal{M}(\mu_X)\) and \(Y \sim \mathcal{M}(\mu_Y)\).
Parameters. The means of the two populations \(\mu_X\) and \(\mu_Y\) are unknown, and what we want to study…
Samples. We assume the two samples \(\mathbf{x} = (x_1, x_2, \ldots, x_n)\) and \(\mathbf{y} = (y_1, y_2, \ldots, y_m)\) are randomly selected from the populations \(X\) and \(Y\).
Estimate (statistic) \(\hat{d} = \overline{\mathbf{x}} - \overline{\mathbf{y}}\): the difference between means of the two samples \(\mathbf{x}\) and \(\mathbf{y}\).
Estimator \(\DMeans\): the function that computes the difference between means from two samples, \(\hat{d} = \DMeans(\mathbf{x},\mathbf{y}) = \overline{\mathbf{x}} - \overline{\mathbf{y}}\). the function we use to compute the estimate \(\hat{d}\).
Sampling distribution \(\hat{D} = \DMeans(\mathbf{X}, \mathbf{Y})\): the difference between means computed from two random samples \(\mathbf{X}\) and \(\mathbf{Y}\).
def mean(sample):
return sum(sample) / len(sample)
def var(sample):
xbar = mean(sample)
sumsqdevs = sum([(xi-xbar)**2 for xi in sample])
return sumsqdevs / (len(sample)-1)
def std(sample):
s2 = var(sample)
return np.sqrt(s2)
def dmeans(xsample, ysample):
dhat = mean(xsample) - mean(ysample)
return dhat
Comparing two groups#
Description of the problem: compare two group based on sample means computed from each group.
Computing \(p\)-values#
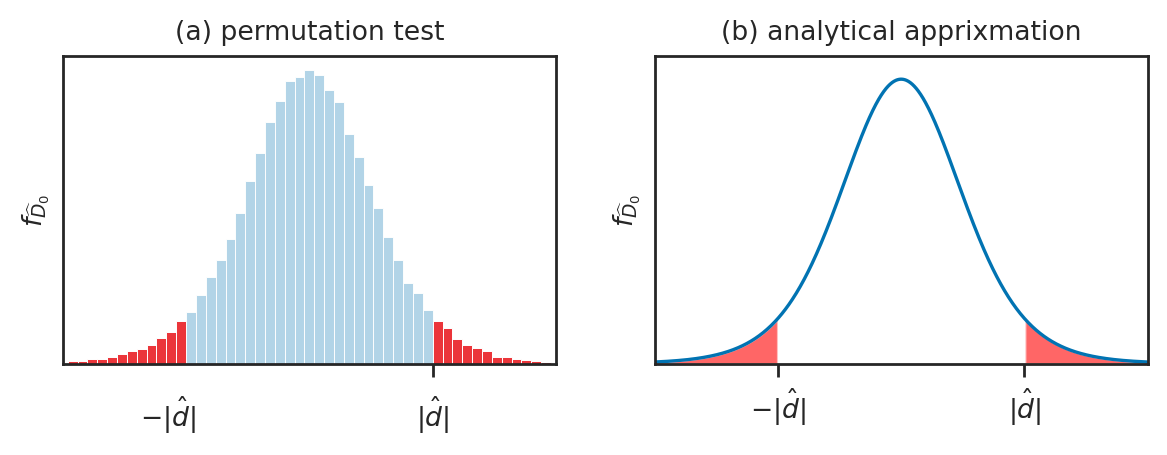
Permutation test for comparing two populations#
The permutations test is a computational approach for comparing two group. The permutation test takes it’s name from the action of mixing up the group-membership labels and computing a statistic which is a way to generate samples from the null hypothesis in situations where the null hypothesis states there is no difference between two groups.
Permutations#
np.random.seed(43)
np.random.permutation([1,2,3,4,5,6,7])
array([4, 6, 3, 7, 2, 1, 5])
np.random.seed(45)
np.random.permutation([1,2,3,4,5,6,7])
array([6, 3, 2, 1, 5, 7, 4])
Resampling under the null hypothesis#
def resample_under_H0(xsample, ysample):
"""
Generate new samples from a random permutation of
the values in the samples `xsample` and `xsample`.
"""
values = np.concatenate((xsample, ysample))
pvalues = np.random.permutation(values)
pxsample = pvalues[0:len(xsample)]
pysample = pvalues[len(xsample):]
return pxsample, pysample
# example 1
np.random.seed(43)
resample_under_H0([1,2,3], [4,5,6,7])
(array([4, 6, 3]), array([7, 2, 1, 5]))
# example 2
np.random.seed(45)
resample_under_H0([1,2,3], [4,5,6,7])
(array([6, 3, 2]), array([1, 5, 7, 4]))
The function resample_under_H0 gives us a way to generate samples from the null hypothesis.
We can then compute the value of the dmeans statistic for these samples. We used the assumption of “no difference” under the null hypothesis, and translated this to the “forget the labels” interpretation.
Example 5P: electricity prices#
The probability model under the alternative hypothesis describes two populations with different parameters:
Under the null hypothesis the populations have the same mean:
eprices = pd.read_csv("datasets/eprices.csv")
eprices
| loc | price | |
|---|---|---|
| 0 | East | 7.7 |
| 1 | East | 5.9 |
| 2 | East | 7.0 |
| 3 | East | 4.8 |
| 4 | East | 6.3 |
| 5 | East | 6.3 |
| 6 | East | 5.5 |
| 7 | East | 5.4 |
| 8 | East | 6.5 |
| 9 | West | 11.8 |
| 10 | West | 10.0 |
| 11 | West | 11.0 |
| 12 | West | 8.6 |
| 13 | West | 8.3 |
| 14 | West | 9.4 |
| 15 | West | 8.0 |
| 16 | West | 6.8 |
| 17 | West | 8.5 |
eprices.groupby("loc")["price"].describe()
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| loc | ||||||||
| East | 9.0 | 6.155556 | 0.877655 | 4.8 | 5.5 | 6.3 | 6.5 | 7.7 |
| West | 9.0 | 9.155556 | 1.562139 | 6.8 | 8.3 | 8.6 | 10.0 | 11.8 |
pricesW = eprices[eprices["loc"]=="West"]["price"]
pricesE = eprices[eprices["loc"]=="East"]["price"]
# 1. Calculate the observed difference between means
dprice = dmeans(pricesW, pricesE)
dprice
3.0
Our goal is to determine how likely or unlikely the observed value \(\hat{d}=3\) is under the null hypothesis \(H_0\). This means we need to obtain the sampling distribution of \(\hat{D}\) under \(H_0\), which we can do using the permutation test.
Let’s look at some of the differences we can expect to observe under \(H_0\).
#######################################################
np.random.seed(31)
# generate new samples by shuffling the labels
ppricesW, ppricesE = resample_under_H0(pricesW, pricesE)
# Compute the difference in means for the bootstrap samples
pdhat0 = dmeans(ppricesW, ppricesE)
pdhat0
1.6444444444444475
Running a permutation test#
We can repeat the resampling procedure 10000 times to get the sampling distribution of \(\widehat{D}_0\) under \(H_0\),
as illustrated in the code procedure below.
np.random.seed(42)
# 2. Obtain the sampling distribution under H0
P = 10000
pdhats0 = []
for i in range(0, P):
ps1, ps2 = resample_under_H0(pricesW, pricesE)
pdhat0 = dmeans(ps1, ps2)
pdhats0.append(pdhat0)
Now that we have the sampling distribution \(\widehat{D}_0\),
we can see where the observed value \(\hat{d}= 3\) =dprice falls within this distribution.
The \(p\)-value is the probability of observing value dprice=3 or more extreme under the null hypothesis.
# 3. Compute the p-value
tails = tailvalues(pdhats0, dprice)
pvalue = len(tails) / len(pdhats0)
pvalue
0.0002
# plot the sampling distribution in blue
bins = nicebins(pdhats0, dprice)
ax = sns.histplot(pdhats0, bins=bins, alpha=0.3)
# plot red line for the observed statistic
plt.axvline(dprice, color="red")
# plot the values that are equal or more extreme in red
sns.histplot(tails, ax=ax, bins=bins, color="red")
ax.set_ylabel(r"$f_{\widehat{D}_0}$");
Effect size estimates#
dprice = dmeans(pricesW, pricesE)
dprice
3.0
The bootstrap confidence interval for the effect size \(\ci{\Delta,0.9}^*\) tells us a range of plausible values for the effect size.
from ministats import ci_dmeans
np.random.seed(45)
ci_dmeans(pricesW, pricesE, alpha=0.1, method="b")
[2.0777777777777775, 3.9555555555555557]
The 90% confidence interval \(\ci{\Delta,0.9}^* = [2.08, 3.96]\) describes an interval of numbers that should contain the difference between group means \(\Delta\) at least 90% of the time:
Reusable function#
def permutation_test_dmeans(xsample, ysample, P=10000):
# 1. Compute the observed difference between means
obsdhat = dmeans(xsample, ysample)
# 2. Get sampling dist. of `dmeans` under H0
pdhats0 = []
for i in range(0, P):
psx, psy = resample_under_H0(xsample, ysample)
pdhat0 = dmeans(psx, psy)
pdhats0.append(pdhat0)
# 3. Compute the p-value
tails = tailvalues(pdhats0, obsdhat, alt="two-sided")
pvalue = len(tails) / len(pdhats0)
return pvalue
np.random.seed(42)
permutation_test_dmeans(pricesW, pricesE)
0.0002
Example 6P: lecture and debate curriculums#
students = pd.read_csv("datasets/students.csv")
students
| student_ID | background | curriculum | effort | score | |
|---|---|---|---|---|---|
| 0 | 1 | arts | debate | 10.96 | 75.0 |
| 1 | 2 | science | lecture | 8.69 | 75.0 |
| 2 | 3 | arts | debate | 8.60 | 67.0 |
| 3 | 4 | arts | lecture | 7.92 | 70.3 |
| 4 | 5 | science | debate | 9.90 | 76.1 |
| 5 | 6 | business | debate | 10.80 | 79.8 |
| 6 | 7 | science | lecture | 7.81 | 72.7 |
| 7 | 8 | business | lecture | 9.13 | 75.4 |
| 8 | 9 | business | lecture | 5.21 | 57.0 |
| 9 | 10 | science | lecture | 7.71 | 69.0 |
| 10 | 11 | business | debate | 9.82 | 70.4 |
| 11 | 12 | arts | debate | 11.53 | 96.2 |
| 12 | 13 | science | debate | 7.10 | 62.9 |
| 13 | 14 | science | lecture | 6.39 | 57.6 |
| 14 | 15 | arts | debate | 12.00 | 84.3 |
students.groupby("curriculum")["score"].describe()
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| curriculum | ||||||||
| debate | 8.0 | 76.462500 | 10.519633 | 62.9 | 69.55 | 75.55 | 80.925 | 96.2 |
| lecture | 7.0 | 68.142857 | 7.758406 | 57.0 | 63.30 | 70.30 | 73.850 | 75.4 |
studentsD = students[students["curriculum"]=="debate"]
studentsL = students[students["curriculum"]=="lecture"]
scoresD = studentsD["score"]
scoresL = studentsL["score"]
# observed difference between score means
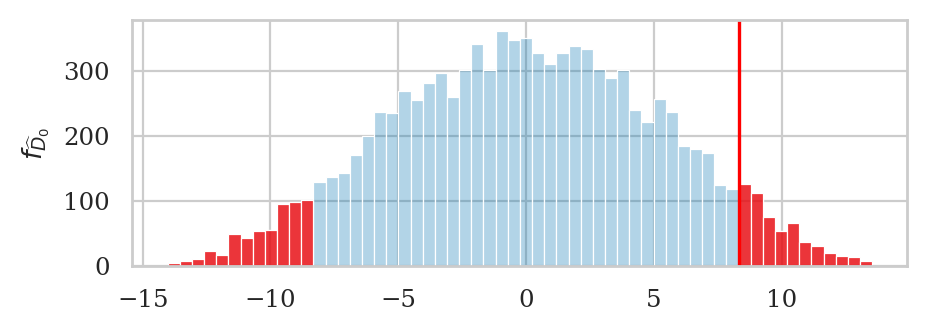
dscore = dmeans(scoresD, scoresL)
dscore
8.319642857142853
Our goal is to determine how likely or unlikely this observed value \(\hat{d}_s=8.32\) is under the null hypothesis \(H_0\). This means we need to obtain the sampling distribution of \(\hat{D}_0\) under \(H_0\), which we can do using the permutation test.
np.random.seed(43)
pvalue = permutation_test_dmeans(scoresD, scoresL)
pvalue
0.1118
# generate sampling distribution under H0
np.random.seed(43)
pdhats = []
P = 10000
for i in range(0, P):
rs1, rs2 = resample_under_H0(scoresD, scoresL)
pdhat = dmeans(rs1, rs2)
pdhats.append(pdhat)
# plot the sampling distribution in blue
bins = nicebins(pdhats, dscore)
ax = sns.histplot(pdhats, bins=bins, alpha=0.3)
# plot red line for the observed statistic
plt.axvline(dscore, color="red")
# plot the values that are equal or more extreme in red
tails = tailvalues(pdhats, dscore)
# print( len(tails) / len(pdhats) )
sns.histplot(tails, bins=bins, ax=ax, color="red")
ax.set_ylabel(r"$f_{\widehat{D}_0}$");
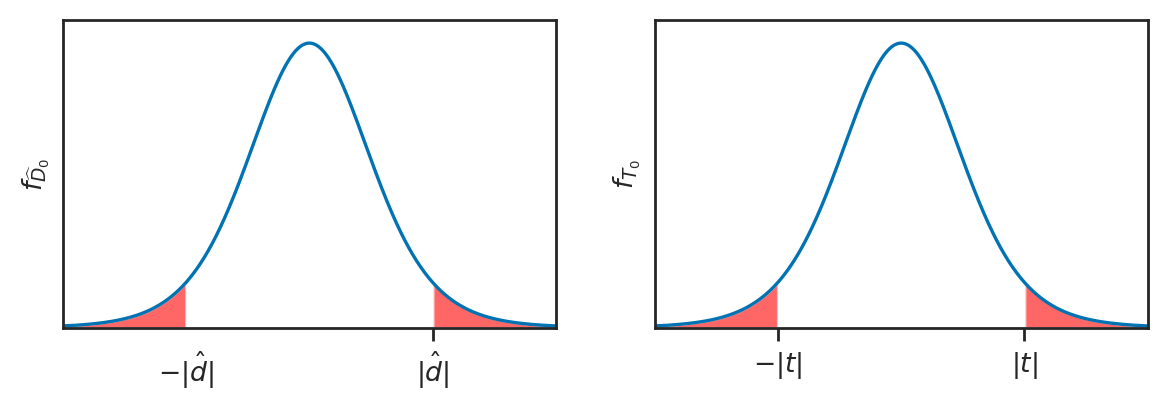
Analytical approximation methods#
We’ll now look at another approach for computing the sampling distribution of the difference between means estimator, using an analytical approximations based on Student’s \(t\)-distribution. This is called the “two-sample \(t\)-test” and it is procedure most commonly taught approach in STATS 101 courses.
How likely or unlikely is the observed difference \(d=3\) under the null hypothesis \(H_0\)?
Analytical approximations are math models for describing the sampling distribution under \(H_0\)
Real sampling distributions: obtained by repeated sampling from \(H_0\)
Analytical approximation: math model based on estimated parameters
Based on this assumption we can use the theoretical model for the difference between group means (we assumed the two unknown populations are normally distributed), we can obtain a closed form expression for the sampling distribution of \(D_0\).
In particular, the probability model for the two groups under \(H_0\) are:
\[ H_0: \qquad X = \mathcal{N}({\color{red}{\mu_0}}, \sigma_X) \quad \textrm{and} \quad Y = \mathcal{N}({\color{red}{\mu_0}}, \sigma_Y), \quad \]from which we can derive the model for \(D = \overline{\mathbf{X}} - \overline{\mathbf{Y}}\):
\[ D_0 \sim \mathcal{N}\!\left( {\color{red}{0}}, \; \stderr{\hat{d}} \right) \]In words, the sampling distribution of the difference between group means is normally distributed with mean \(\Delta = 0\) and standard deviation \(\stderrhat{\hat{d}}\), described by the formula:
\[ \stderr{\hat{d}} % \; = \; \sqrt{ \stderr{\overline{\mathbf{x}}}^2 + \stderr{\overline{\mathbf{y}}}^2 } = \sqrt{ \tfrac{\sigma_X^2}{n} + \tfrac{\sigma_Y^2}{m} }\;. \]The standard error of the difference between means depends on the variance of the two groups \(\sigma_X^2\) and \(\sigma_Y^2\). Recall we’ve seen this expression earlier in Section 3.1 and Section 3.2 as well.
Problem 1: the population variances \(\sigma_X^2\) and \(\sigma_Y^2\) are unknown, and we only have the estimated variances \(s_{\mathbf{x}}\) and \(s_{\mathbf{y}}\), which we calculated from the samples \(\mathbf{x}\) and \(\mathbf{y}\).
Solution 1: use plug in estimate and We can obtain an approximation to the standard error of the difference between means estimator using the plug-in principle, which states we can use search-and-replace the population parameters \(\sigma_X^2\) and \(\sigma_Y^2\) with the estimates \(s_{\mathbf{x}}\) and \(s_{\mathbf{y}}\):
\[ \stderrhat{\hat{d}} % \; = \; \sqrt{ \stderrhat{\overline{\mathbf{x}}}^2 + \stderrhat{\overline{\mathbf{y}}}^2 } = \sqrt{ \tfrac{s_{\mathbf{x}}^2}{n} + \tfrac{s_{\mathbf{y}}^2}{m} }\;. \]Problem 2: the estimated standard error \(\stderrhat{\hat{d}}\) tends to underestimate the true standard error \(\stderr{\hat{d}}\), so when we use the basic normal model \(\mathcal{N}\!\left(\tt{loc}=0, \tt{scale}=\stderrhat{\hat{d}} \right)\) the approximation is off (not enough weight in the tails).
Solution 2: use Student’s \(t\)-distribution. We can define a new model for the \(D_0 = \overline{\mathbf{X}} - \overline{\mathbf{Y}}\) based on Student’s \(t\)-distribution: $\( D_0 \sim \mathcal{T}\!\left(\tt{df}=\nu_d, \; \tt{loc}={\color{red}{0}}, \; \tt{scale}=\stderrhat{\hat{d}} \right) \)\( In words, the sampling distribution of the difference between group means can be modeled as Student's \)t\(-distribution with \)\nu_d\( degrees of freedom, centered at \)0\(, with scale parameter \)\stderrhat{\hat{d}}$.
The degrees of freedom parameter, denoted \(\nu_d\) (Greek letter nu) in equations (
dfin code) determines the “weight” in the tails of the distribution. It is computed by a complicated formula that depends on the sample sizes \(n\) and \(m\), and the sample standard deviations \(s_{\mathbf{x}}\) and \(s_{\mathbf{y}}\).
Example 5T: comparing East and West electricity prices#
eprices = pd.read_csv("datasets/eprices.csv")
pricesW = eprices[eprices["loc"]=="West"]["price"]
pricesE = eprices[eprices["loc"]=="East"]["price"]
# Calculate the observed difference between means
dprice = dmeans(pricesW, pricesE)
dprice
3.0
Two-sample t-test#
We’ll now show the data, probability modeling, and calculation steps of the “two-sample \(t\)-test for comparing group means. The two-sample \(t\)-test is also known as Welch’s t-test.
# Calculate the sample size and the standard deviation for each group
nW, nE = len(pricesW), len(pricesE)
stdW, stdE = std(pricesW), std(pricesE)
# Compute the standard error of the estimator D
seD = np.sqrt(stdW**2/nW + stdE**2/nE)
seD
0.5972674401486561
# Compute the value of the t-statistic
obst = (dprice - 0) / seD
obst
5.022875513276463
from ministats import calcdf
# Obtain the degrees of freedom from the crazy formula
dfD = calcdf(stdW, nW, stdE, nE)
dfD
12.59281702723103
# Calculate the p-value
from scipy.stats import t as tdist
rvT0 = tdist(df=dfD)
from ministats import tailprobs
pvalue = tailprobs(rvT0, obst, alt="two-sided")
pvalue
0.00025703383372176864
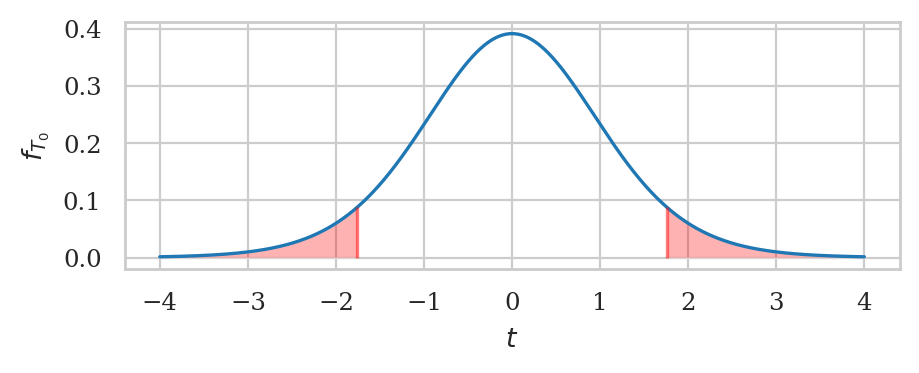
# # SKIP BECAUSE TOO SMALL TO SEE
# _, ax = calc_prob_and_plot_tails(rvT0, -obst, obst, xlims=[-6,6])
# ax.set_title(None)
# ax.set_xlabel("$t$")
# ax.set_ylabel("$f_{T_0}$");
Effect size estimates#
dprice = dmeans(pricesW, pricesE)
dprice
3.0
The confidence interval for the effect size \(\ci{\Delta,0.9}\) tells us a range of plausible values for the effect size.
from ministats import ci_dmeans
ci_dmeans(pricesW, pricesE, alpha=0.1, method="a")
[1.9396575883681466, 4.060342411631854]
The 90% confidence interval \(\ci{\Delta,0.9} = [1.94, 4.06]\) describes an interval of numbers that should contain the difference between group means \(\Delta\) at least 90% of the time:
Reusable procedure for two-sample \(t\)-test#
def ttest_dmeans(xsample, ysample):
# Calculate the observed difference between means
obsdhat = mean(xsample) - mean(ysample)
# Calculate the sample sizes and the stds
n, m = len(xsample), len(ysample)
sx, sy = std(xsample), std(ysample)
# Calculate the standard error, the degrees of
# freedom, the null model, and the t-statistic
seD = np.sqrt(sx**2/n + sy**2/m)
dfD = calcdf(sx, n, sy, m)
obst = (obsdhat - 0) / seD
# Calculate the p-value from the t-distribution
rvT0 = tdist(df=dfD)
pvalue = tailprobs(rvT0, obst, alt="two-sided")
return pvalue
Verify we get the same \(p\)-value when using the standard error formula.
ttest_dmeans(pricesW, pricesE)
0.00025703383372176864
Example 6T: comparing rural and urban sleep scores#
students = pd.read_csv("datasets/students.csv")
# students
# students.groupby("curriculum")["score"].describe()
scoresD = students[students["curriculum"]=="debate"]["score"]
scoresL = students[students["curriculum"]=="lecture"]["score"]
# observed difference between score means
dscores = dmeans(scoresD, scoresL)
dscores
8.319642857142853
ttest_dmeans(scoresD, scoresL)
0.10307495448387446
# T-TEST ANALYSIS
nD, nL = len(scoresD), len(scoresL)
stdD, stdL = std(scoresD), std(scoresL)
seDDL = np.sqrt(stdD**2/nD + stdL**2/nL)
obstDL = (dscores - 0) / seDDL
from ministats import calcdf
dfDL = calcdf(stdD, nD, stdL, nL)
rvT0DL = tdist(df=dfDL)
pvalueDL = tailprobs(rvT0DL, obstDL, alt="two-sided")
pvalueDL
0.10307495448387446
Alternative calculation methods#
Two-sample \(t\)-test using pooled variance#
Pooled variance formulas#
Example 5T revisited with pooled variance#
# TODO make self contained
# Compute the pooled variance and standard error of estimator D
varp = ((nW-1)*stdW**2 + (nE-1)*stdE**2) / (nW + nE - 2)
stdp = np.sqrt(varp)
seDp = stdp * np.sqrt(1/nW + 1/nE)
seDp
0.5972674401486561
# Compute the value of the t-statistic
obstp = (dprice - 0) / seDp
obstp
5.022875513276463
# Obtain the degrees of freedom
dfDp = nW + nE - 2
dfDp
16
from scipy.stats import t as tdist
from ministats import tailprobs
# Calculate the p-value
rvT0p = tdist(df=dfDp)
pvaluep = tailprobs(rvT0p, obstp, alt="two-sided")
pvaluep
0.00012497067987674894
Using scipy.stats.ttest_ind for two-sample \(t\)-tests#
Alternatively,
we can use scipy.stats.ttest_ind to obtain the \(p\)-value.
Use equal_var=False to obtain the Welsh value.
# ALT. compute using existing function in `scipy.stats`
from scipy.stats import ttest_ind
res1 = ttest_ind(pricesW, pricesE, equal_var=False)
res1.pvalue
0.0002570338337217609
Use equal_var=True to obtain the pooled variance value.
# ALT. compute using existing function in `scipy.stats`
from scipy.stats import ttest_ind
res2 = ttest_ind(pricesW, pricesE, equal_var=True)
res2.pvalue
0.00012497067987678488
Using scipy.stats.ttest_ind for permutation test#
You can use the SciPy implementation of permutation test,
by calling ttest_ind(..., permutations=10000) to perform a permutation test, then obtain the \(p\)-value.
from scipy.stats import ttest_ind
np.random.seed(42)
ttest_ind(pricesW, pricesE, permutations=10000).pvalue
/tmp/ipykernel_9822/472059880.py:4: DeprecationWarning: Arguments {'permutations'} are deprecated, whether passed by position or keyword. They will be removed in SciPy 1.17.0. Use ``method`` to perform a permutation test.
ttest_ind(pricesW, pricesE, permutations=10000).pvalue
0.00029997000299970003
Note the \(p\)-value we obtained form the two methods may be different. This can happen when using the permutations test, because we use randomness as part of the calculation.
Using statsmodels for two-sample \(t\)-tests#
import statsmodels.stats.api as sms
statsW = sms.DescrStatsW(pricesW)
statsE = sms.DescrStatsW(pricesE)
cm = sms.CompareMeans(statsW, statsE)
# Welch's t-test
cm.ttest_ind(alternative="two-sided", usevar="unequal")
(5.022875513276463, 0.000257033833721762, 12.592817027231028)
# T-test with pooled variance
cm.ttest_ind(alternative="two-sided", usevar="pooled")
(5.022875513276463, 0.00012497067987678534, 16.0)
Explanations#
Statistical modelling assumptions#
INDEP:
NORMAL:
LARGEn:
EQVAR:
Standardized effect size#
It is sometimes useful to report the effect size using a “standardized” measure for effect sizes.
Cohen’s \(d\) is one such measure, and it is defined as the difference between two means divided by the pooled standard deviation.
where the pooled variance is defined as \(s_{p}^2 = [(n -1)s_{\mathbf{x}}^2 + (m -1)s_{\mathbf{y}}^2]/(n + m -2)\).
def cohend2(sample1, sample2):
"""
Cohen's d measure of effect size for two independent samples.
"""
n1, n2 = len(sample1), len(sample2)
mean1, mean2 = mean(sample1), mean(sample2)
var1, var2 = var(sample1), var(sample2)
# calculate the pooled variance and std
var_pooled = ((n1-1)*var1+(n2-1)*var2) / (n1+n2-2)
std_pooled = np.sqrt(var_pooled)
d = (mean1 - mean2) / std_pooled
return d
We interpret the value of Cohen’s \(d\) using the reference table of values:
Cohen’s d |
Effect size |
|---|---|
0.01 |
very small |
0.20 |
small |
0.50 |
medium |
0.80 |
large |
Effect size for mean deviation of kombucha Batch 04 from expected value \(\mu_K=1000\).
cohend2(pricesW, pricesE)
2.3678062243290983
Discussion#
Comparison to resampling methods#
Exercises#
See notebook exercises_35_two_sample_tests.ipynb.